Transcriptomes of the major human pancreatic cell types
- PMID: 21882062
- PMCID: PMC3880150
- DOI: 10.1007/s00125-011-2283-5
Transcriptomes of the major human pancreatic cell types
Erratum in
- Diabetologia. 2013 May;56(5):1192
Abstract
Aims/hypothesis: We sought to determine the mRNA transcriptome of all major human pancreatic endocrine and exocrine cell subtypes, including human alpha, beta, duct and acinar cells. In addition, we identified the cell type-specific distribution of transcription factors, signalling ligands and their receptors.
Methods: Islet samples from healthy human donors were enzymatically dispersed to single cells and labelled with cell type-specific surface-reactive antibodies. Live endocrine and exocrine cell subpopulations were isolated by FACS and gene expression analyses were performed using microarray analysis and quantitative RT-PCR. Computational tools were used to evaluate receptor-ligand representation in these populations.
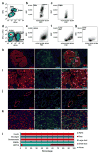
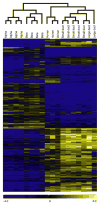
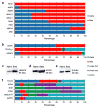
Results: Analysis of the transcriptomes of alpha, beta, large duct, small duct and acinar cells revealed previously unrecognised gene expression patterns in these cell types, including transcriptional regulators HOPX and HDAC9 in the human beta cell population. The abundance of some regulatory proteins was different from that reported in mouse tissue. For example, v-maf musculoaponeurotic fibrosarcoma oncogene homologue B (avian) (MAFB) was detected at equal levels in adult human alpha and beta cells, but is absent from adult mouse beta cells. Analysis of ligand-receptor interactions suggested that EPH receptor-ephrin communication between exocrine and endocrine cells contributes to pancreatic function.
Conclusions/interpretation: This is the first comprehensive analysis of the transcriptomes of human exocrine and endocrine pancreatic cell types-including beta cells-and provides a useful resource for diabetes research. In addition, paracrine signalling pathways within the pancreas are shown. These results will help guide efforts to specify human beta cell fate by embryonic stem cell or induced pluripotent stem cell differentiation or genetic reprogramming.
Figures
Similar articles
-
Transcriptome analysis of pancreatic cells across distant species highlights novel important regulator genes.BMC Biol. 2017 Mar 21;15(1):21. doi: 10.1186/s12915-017-0362-x. BMC Biol. 2017. PMID: 28327131 Free PMC article.
-
Thyroid hormones promote endocrine differentiation at expenses of exocrine tissue.Exp Cell Res. 2014 Apr 1;322(2):236-48. doi: 10.1016/j.yexcr.2014.01.030. Epub 2014 Feb 4. Exp Cell Res. 2014. PMID: 24503054
-
Specific control of pancreatic endocrine β- and δ-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9.Diabetes. 2011 Nov;60(11):2861-71. doi: 10.2337/db11-0440. Epub 2011 Sep 27. Diabetes. 2011. PMID: 21953612 Free PMC article.
-
PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration.Stem Cell Res Ther. 2017 Nov 2;8(1):240. doi: 10.1186/s13287-017-0694-z. Stem Cell Res Ther. 2017. PMID: 29096722 Free PMC article. Review.
-
Multiple functions of Maf in the regulation of cellular development and differentiation.Diabetes Metab Res Rev. 2015 Nov;31(8):773-8. doi: 10.1002/dmrr.2676. Epub 2015 Aug 18. Diabetes Metab Res Rev. 2015. PMID: 26122665 Free PMC article. Review.
Cited by
-
α-cell role in β-cell generation and regeneration.Islets. 2012 May-Jun;4(3):188-98. doi: 10.4161/isl.20500. Islets. 2012. PMID: 22847495 Free PMC article. Review.
-
Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice.Nat Commun. 2016 Aug 30;7:12631. doi: 10.1038/ncomms12631. Nat Commun. 2016. PMID: 27572106 Free PMC article.
-
Directed differentiation of cholangiocytes from human pluripotent stem cells.Nat Biotechnol. 2015 Aug;33(8):853-61. doi: 10.1038/nbt.3294. Epub 2015 Jul 13. Nat Biotechnol. 2015. PMID: 26167630
-
DNA methylation reveals distinct cells of origin for pancreatic neuroendocrine carcinomas and pancreatic neuroendocrine tumors.Genome Med. 2022 Mar 1;14(1):24. doi: 10.1186/s13073-022-01018-w. Genome Med. 2022. PMID: 35227293 Free PMC article.
-
Metabolic regulation through the endosomal system.Traffic. 2019 Aug;20(8):552-570. doi: 10.1111/tra.12670. Epub 2019 Jun 24. Traffic. 2019. PMID: 31177593 Free PMC article. Review.
References
-
- Maffei A, Liu Z, Witkowski P, et al. Identification of tissue-restricted transcripts in human islets. Endocrinology. 2004;145:4513–4521. - PubMed
-
- Gunton JE, Kulkarni RN, Yim S, et al. Loss of ARNT/HIF1β mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–349. - PubMed
-
- Lyttle BM, Li J, Krishnamurthy M, et al. Transcription factor expression in the developing human fetal endocrine pancreas. Diabetologia. 2008;51:1169–1180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

