Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks
- PMID: 21880593
- PMCID: PMC3239177
- DOI: 10.1093/nar/gkr656
Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks
Abstract
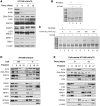
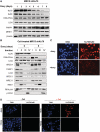
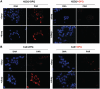
In mammalian cells, the main pathway for DNA double-strand breaks (DSBs) repair is classical non-homologous end joining (C-NHEJ). An alternative or back-up NHEJ (B-NHEJ) pathway has emerged which operates preferentially under C-NHEJ defective conditions. Although B-NHEJ appears particularly relevant to genomic instability associated with cancer, its components and regulation are still largely unknown. To get insights into this pathway, we have knocked-down Ku, the main contributor to C-NHEJ. Thus, models of human cell lines have been engineered in which the expression of Ku70/80 heterodimer can be significantly lowered by the conditional induction of a shRNA against Ku70. On Ku reduction in cells, resulting NHEJ competent protein extracts showed a shift from C- to B-NHEJ that could be reversed by addition of purified Ku protein. Using a cellular fractionation protocol after treatment with a strong DSBs inducer followed by western blotting or immunostaining, we established that, among C-NHEJ factors, Ku is the main counteracting factor against mobilization of PARP1 and the MRN complex to damaged chromatin. In addition, Ku limits PAR synthesis and single-stranded DNA production in response to DSBs. These data support the involvement of PARP1 and the MRN proteins in the B-NHEJ route for the repair of DNA DSBs.
© The Author(s) 2011. Published by Oxford University Press.
Figures




Similar articles
-
The absence of Ku but not defects in classical non-homologous end-joining is required to trigger PARP1-dependent end-joining.DNA Repair (Amst). 2013 Dec;12(12):1134-42. doi: 10.1016/j.dnarep.2013.10.005. Epub 2013 Nov 7. DNA Repair (Amst). 2013. PMID: 24210699
-
PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways.Nucleic Acids Res. 2006;34(21):6170-82. doi: 10.1093/nar/gkl840. Epub 2006 Nov 6. Nucleic Acids Res. 2006. PMID: 17088286 Free PMC article.
-
Histone deacetylase inhibitors decrease NHEJ both by acetylation of repair factors and trapping of PARP1 at DNA double-strand breaks in chromatin.Leuk Res. 2016 Jun;45:14-23. doi: 10.1016/j.leukres.2016.03.007. Epub 2016 Mar 30. Leuk Res. 2016. PMID: 27064363 Free PMC article.
-
The Ku heterodimer: function in DNA repair and beyond.Mutat Res Rev Mutat Res. 2015 Jan-Mar;763:15-29. doi: 10.1016/j.mrrev.2014.06.002. Epub 2014 Jul 4. Mutat Res Rev Mutat Res. 2015. PMID: 25795113 Review.
-
One ring to bring them all--the role of Ku in mammalian non-homologous end joining.DNA Repair (Amst). 2014 May;17:30-8. doi: 10.1016/j.dnarep.2014.02.019. Epub 2014 Mar 26. DNA Repair (Amst). 2014. PMID: 24680220 Review.
Cited by
-
CRISPR-based gene editing of non-homologous end joining factors biases DNA repair pathway choice toward single-strand annealing in Aedes aegypti.Curr Res Biotechnol. 2023;5:100133. doi: 10.1016/j.crbiot.2023.100133. Epub 2023 May 29. Curr Res Biotechnol. 2023. PMID: 37475832 Free PMC article.
-
PARG is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(ADP-ribose) upon prolonged replicative stress.Nucleic Acids Res. 2014 Jul;42(12):7776-92. doi: 10.1093/nar/gku505. Epub 2014 Jun 6. Nucleic Acids Res. 2014. PMID: 24906880 Free PMC article.
-
Balancing self-renewal against genome preservation in stem cells: How do they manage to have the cake and eat it too?Cell Mol Life Sci. 2016 May;73(9):1803-23. doi: 10.1007/s00018-016-2152-y. Epub 2016 Feb 17. Cell Mol Life Sci. 2016. PMID: 26886024 Free PMC article. Review.
-
XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.Nat Struct Mol Biol. 2018 Oct;25(10):971-980. doi: 10.1038/s41594-018-0133-6. Epub 2018 Oct 5. Nat Struct Mol Biol. 2018. PMID: 30291363 Free PMC article.
-
PARP-1: a critical regulator in radioprotection and radiotherapy-mechanisms, challenges, and therapeutic opportunities.Front Pharmacol. 2023 Jun 6;14:1198948. doi: 10.3389/fphar.2023.1198948. eCollection 2023. Front Pharmacol. 2023. PMID: 37351512 Free PMC article. Review.
References
-
- O'Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat. Rev. Genet. 2006;7:45–54. - PubMed
-
- Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 2006;40:363–383. - PubMed
-
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

