The first two nucleotides of the respiratory syncytial virus antigenome RNA replication product can be selected independently of the promoter terminus
- PMID: 21878549
- PMCID: PMC3185921
- DOI: 10.1261/rna.2813411
The first two nucleotides of the respiratory syncytial virus antigenome RNA replication product can be selected independently of the promoter terminus
Abstract
There is limited knowledge regarding how the RNA-dependent RNA polymerases of the nonsegmented negative-strand RNA viruses initiate genome replication. In a previous study of respiratory syncytial virus (RSV) RNA replication, we found evidence that the polymerase could select the 5'-ATP residue of the genome RNA independently of the 3' nucleotide of the template. To investigate if a similar mechanism is used during antigenome synthesis, a study of initiation from the RSV leader (Le) promoter was performed using an intracellular minigenome assay in which RNA replication was restricted to a single step, so that the products examined were derived only from input mutant templates. Templates in which Le nucleotides 1U, or 1U and 2G, were deleted directed efficient replication, and in both cases, the replication products were initiated at the wild-type position, at position -1 or -2 relative to the template, respectively. Sequence analysis of the RNA products showed that they contained ATP and CTP at the -1 and -2 positions, respectively, thus restoring the mini-antigenome RNA to wild-type sequence. These data indicate that the RSV polymerase is able to select the first two nucleotides of the antigenome and initiate at the correct position, even if the 3'-terminal two nucleotides of the template are missing. Substitution of positions +1 and +2 of the template reduced RNA replication and resulted in increased initiation at positions +3 and +5. Together these data suggest a model for how the RSV polymerase initiates antigenome synthesis.
Figures

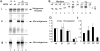
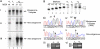
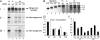

Similar articles
-
Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion.Proc Natl Acad Sci U S A. 2010 Jun 1;107(22):10226-31. doi: 10.1073/pnas.0913065107. Epub 2010 May 17. Proc Natl Acad Sci U S A. 2010. PMID: 20479224 Free PMC article.
-
Evidence that the respiratory syncytial virus polymerase is recruited to nucleotides 1 to 11 at the 3' end of the nucleocapsid and can scan to access internal signals.J Virol. 2005 Sep;79(17):11311-22. doi: 10.1128/JVI.79.17.11311-11322.2005. J Virol. 2005. PMID: 16103183 Free PMC article.
-
Mutations in the 5' trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step.J Virol. 2000 Jan;74(1):146-55. doi: 10.1128/jvi.74.1.146-155.2000. J Virol. 2000. PMID: 10590101 Free PMC article.
-
Unravelling the complexities of respiratory syncytial virus RNA synthesis.J Gen Virol. 2006 Jul;87(Pt 7):1805-1821. doi: 10.1099/vir.0.81786-0. J Gen Virol. 2006. PMID: 16760383 Review.
-
Structural Insights into the Respiratory Syncytial Virus RNA Synthesis Complexes.Viruses. 2021 May 5;13(5):834. doi: 10.3390/v13050834. Viruses. 2021. PMID: 34063087 Free PMC article. Review.
Cited by
-
Biochemistry of the Respiratory Syncytial Virus L Protein Embedding RNA Polymerase and Capping Activities.Viruses. 2023 Jan 25;15(2):341. doi: 10.3390/v15020341. Viruses. 2023. PMID: 36851554 Free PMC article. Review.
-
Complete genome sequences of one human respiratory syncytial antigenic group a virus from china and its four mouse-adapted isolates.Genome Announc. 2015 Mar 5;3(2):e00062-15. doi: 10.1128/genomeA.00062-15. Genome Announc. 2015. PMID: 25744999 Free PMC article.
-
Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease.Curr Top Microbiol Immunol. 2013;372:3-38. doi: 10.1007/978-3-642-38919-1_1. Curr Top Microbiol Immunol. 2013. PMID: 24362682 Free PMC article.
-
Mechanism for de novo initiation at two sites in the respiratory syncytial virus promoter.Nucleic Acids Res. 2018 Jul 27;46(13):6785-6796. doi: 10.1093/nar/gky480. Nucleic Acids Res. 2018. PMID: 29873775 Free PMC article.
-
Respiratory syncytial virus M2-1 protein associates non-specifically with viral messenger RNA and with specific cellular messenger RNA transcripts.PLoS Pathog. 2021 May 18;17(5):e1009589. doi: 10.1371/journal.ppat.1009589. eCollection 2021 May. PLoS Pathog. 2021. PMID: 34003848 Free PMC article.
References
-
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW 2006. Crystal structure of the rabies virus nucleoprotein–RNA complex. Science 313: 360–363 - PubMed
-
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410: 235–240 - PubMed
-
- Chetverin AB, Chetverina HV, Munishkin AV 1991. On the nature of spontaneous RNA synthesis by Qβ replicase. J Mol Biol 222: 3–9 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
