The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate
- PMID: 21878548
- PMCID: PMC3185919
- DOI: 10.1261/rna.2880311
The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate
Abstract
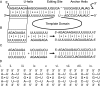
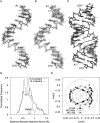
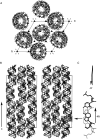
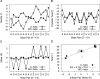
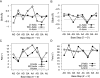
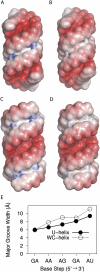
Guide RNAs bind antiparallel to their target pre-mRNAs to form editing substrates in reaction cycles that insert or delete uridylates (Us) in most mitochondrial transcripts of trypanosomes. The 5' end of each guide RNA has an anchor sequence that binds to the pre-mRNA by base-pair complementarity. The template sequence in the middle of the guide RNA directs the editing reactions. The 3' ends of most guide RNAs have ∼15 contiguous Us that bind to the purine-rich unedited pre-mRNA upstream of the editing site. The resulting U-helix is rich in G·U wobble base pairs. To gain insights into the structure of the U-helix, we crystallized 8 bp of the U-helix in one editing substrate for the A6 mRNA of Trypanosoma brucei. The fragment provides three samples of the 5'-AGA-3'/5'-UUU-3' base-pair triple. The fusion of two identical U-helices head-to-head promoted crystallization. We obtained X-ray diffraction data with a resolution limit of 1.37 Å. The U-helix had low and high twist angles before and after each G·U wobble base pair; this variation was partly due to shearing of the wobble base pairs as revealed in comparisons with a crystal structure of a 16-nt RNA with all Watson-Crick base pairs. Both crystal structures had wider major grooves at the junction between the poly(U) and polypurine tracts. This junction mimics the junction between the template helix and the U-helix in RNA-editing substrates and may be a site of major groove invasion by RNA editing proteins.
Figures

Similar articles
-
RNA editing in Trypanosoma brucei: characterization of gRNA U-tail interactions with partially edited mRNA substrates.Nucleic Acids Res. 2001 Feb 1;29(3):703-9. doi: 10.1093/nar/29.3.703. Nucleic Acids Res. 2001. PMID: 11160892 Free PMC article.
-
Structural studies of a double-stranded RNA from trypanosome RNA editing by small-angle X-ray scattering.Methods Mol Biol. 2015;1240:165-89. doi: 10.1007/978-1-4939-1896-6_13. Methods Mol Biol. 2015. PMID: 25352145
-
Fusion RNAs in crystallographic studies of double-stranded RNA from trypanosome RNA editing.Methods Mol Biol. 2015;1240:191-216. doi: 10.1007/978-1-4939-1896-6_14. Methods Mol Biol. 2015. PMID: 25352146
-
RNA editing in trypanosomes.Eur J Biochem. 1994 Apr 1;221(1):9-23. doi: 10.1111/j.1432-1033.1994.tb18710.x. Eur J Biochem. 1994. PMID: 7513284 Review.
-
An innate twist between Crick's wobble and Watson-Crick base pairs.RNA. 2013 Aug;19(8):1038-53. doi: 10.1261/rna.036905.112. RNA. 2013. PMID: 23861536 Free PMC article. Review.
Cited by
-
Direct-methods structure determination of a trypanosome RNA-editing substrate fragment with translational pseudosymmetry.Acta Crystallogr D Struct Biol. 2016 Apr;72(Pt 4):477-87. doi: 10.1107/S2059798316001224. Epub 2016 Mar 24. Acta Crystallogr D Struct Biol. 2016. PMID: 27050127 Free PMC article.
-
Engineering Crystal Packing in RNA Structures I: Past and Future Strategies for Engineering RNA Packing in Crystals.Crystals (Basel). 2021 Aug;11(8):952. doi: 10.3390/cryst11080952. Epub 2021 Aug 15. Crystals (Basel). 2021. PMID: 34745656 Free PMC article.
-
Targeting RNA Structure to Inhibit Editing in Trypanosomes.Int J Mol Sci. 2023 Jun 14;24(12):10110. doi: 10.3390/ijms241210110. Int J Mol Sci. 2023. PMID: 37373258 Free PMC article.
-
Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.Nucleic Acids Res. 2021 Jan 11;49(1):568-583. doi: 10.1093/nar/gkaa1197. Nucleic Acids Res. 2021. PMID: 33332555 Free PMC article.
-
Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters.Biochemistry. 2012 Apr 24;51(16):3508-22. doi: 10.1021/bi3002709. Epub 2012 Apr 10. Biochemistry. 2012. PMID: 22490167 Free PMC article.
References
-
- Arnott S, Hukins DW, Dover SD 1972. Optimised parameters for RNA double-helices. Biochem Biophys Res Commun 48: 1392–1399 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
