Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites
- PMID: 21862836
- PMCID: PMC3195571
- DOI: 10.1074/jbc.C111.284620
Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites
Abstract
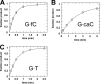
Thymine DNA glycosylase (TDG) excises T from G·T mispairs and is thought to initiate base excision repair (BER) of deaminated 5-methylcytosine (mC). Recent studies show that TDG, including its glycosylase activity, is essential for active DNA demethylation and embryonic development. These and other findings suggest that active demethylation could involve mC deamination by a deaminase, giving a G·T mispair followed by TDG-initiated BER. An alternative proposal is that demethylation could involve iterative oxidation of mC to 5-hydroxymethylcytosine (hmC) and then to 5-formylcytosine (fC) and 5-carboxylcytosine (caC), mediated by a Tet (ten eleven translocation) enzyme, with conversion of caC to C by a putative decarboxylase. Our previous studies suggest that TDG could excise fC and caC from DNA, which could provide another potential demethylation mechanism. We show here that TDG rapidly removes fC, with higher activity than for G·T mispairs, and has substantial caC excision activity, yet it cannot remove hmC. TDG excision of fC and caC, oxidation products of mC, is consistent with its strong specificity for excising bases from a CpG context. Our findings reveal a remarkable new aspect of specificity for TDG, inform its catalytic mechanism, and suggest that TDG could protect against fC-induced mutagenesis. The results also suggest a new potential mechanism for active DNA demethylation, involving TDG excision of Tet-produced fC (or caC) and subsequent BER. Such a mechanism obviates the need for a decarboxylase and is consistent with findings that TDG glycosylase activity is essential for active demethylation and embryonic development, as are mechanisms involving TDG excision of deaminated mC or hmC.
Figures

Similar articles
-
TET-TDG Active DNA Demethylation at CpG and Non-CpG Sites.J Mol Biol. 2021 Apr 16;433(8):166877. doi: 10.1016/j.jmb.2021.166877. Epub 2021 Feb 7. J Mol Biol. 2021. PMID: 33561435 Free PMC article.
-
Divergent mechanisms for enzymatic excision of 5-formylcytosine and 5-carboxylcytosine from DNA.J Am Chem Soc. 2013 Oct 23;135(42):15813-22. doi: 10.1021/ja406444x. Epub 2013 Oct 7. J Am Chem Soc. 2013. PMID: 24063363 Free PMC article.
-
Excision of 5-Carboxylcytosine by Thymine DNA Glycosylase.J Am Chem Soc. 2019 Nov 27;141(47):18851-18861. doi: 10.1021/jacs.9b10376. Epub 2019 Nov 18. J Am Chem Soc. 2019. PMID: 31693361 Free PMC article.
-
Epigenetic modifications in DNA could mimic oxidative DNA damage: A double-edged sword.DNA Repair (Amst). 2015 Aug;32:52-57. doi: 10.1016/j.dnarep.2015.04.013. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956859 Review.
-
Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites.DNA Repair (Amst). 2015 Aug;32:33-42. doi: 10.1016/j.dnarep.2015.04.011. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 26021671 Free PMC article. Review.
Cited by
-
Molecular epigenetic switches in neurodevelopment in health and disease.Front Behav Neurosci. 2015 May 13;9:120. doi: 10.3389/fnbeh.2015.00120. eCollection 2015. Front Behav Neurosci. 2015. PMID: 26029068 Free PMC article. Review.
-
miR-29 represses the activities of DNA methyltransferases and DNA demethylases.Int J Mol Sci. 2013 Jul 12;14(7):14647-58. doi: 10.3390/ijms140714647. Int J Mol Sci. 2013. PMID: 23857059 Free PMC article.
-
Genomic Uracil and Aberrant Profile of Demethylation Intermediates in Epigenetics and Hematologic Malignancies.Int J Mol Sci. 2021 Apr 19;22(8):4212. doi: 10.3390/ijms22084212. Int J Mol Sci. 2021. PMID: 33921666 Free PMC article. Review.
-
Domain Structure of the Dnmt1, Dnmt3a, and Dnmt3b DNA Methyltransferases.Adv Exp Med Biol. 2022;1389:45-68. doi: 10.1007/978-3-031-11454-0_3. Adv Exp Med Biol. 2022. PMID: 36350506 Free PMC article.
-
5-Hydroxymethyl-, 5-Formyl- and 5-Carboxydeoxycytidines as Oxidative Lesions and Epigenetic Marks.Chemistry. 2021 Jun 1;27(31):8100-8104. doi: 10.1002/chem.202100551. Epub 2021 May 1. Chemistry. 2021. PMID: 33769637 Free PMC article.
References
-
- Morgan H. D., Dean W., Coker H. A., Reik W., Petersen-Mahrt S. K. (2004) J. Biol. Chem. 279, 52353–52360 - PubMed
-
- Métivier R., Gallais R., Tiffoche C., Le Péron C., Jurkowska R. Z., Carmouche R. P., Ibberson D., Barath P., Demay F., Reid G., Benes V., Jeltsch A., Gannon F., Salbert G. (2008) Nature 452, 45–50 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

