Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development
- PMID: 21816367
- PMCID: PMC3154739
- DOI: 10.1016/j.stem.2011.07.010
Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development
Abstract
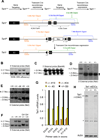
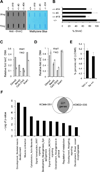
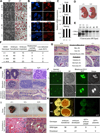
The Tet family of enzymes (Tet1/2/3) converts 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). Mouse embryonic stem cells (mESCs) highly express Tet1 and have an elevated level of 5hmC. Tet1 has been implicated in ESC maintenance and lineage specification in vitro but its precise function in development is not well defined. To establish the role of Tet1 in pluripotency and development, we have generated Tet1 mutant mESCs and mice. Tet1(-/-) ESCs have reduced levels of 5hmC and subtle changes in global gene expression, and are pluripotent and support development of live-born mice in tetraploid complementation assay, but display skewed differentiation toward trophectoderm in vitro. Tet1 mutant mice are viable, fertile, and grossly normal, though some mutant mice have a slightly smaller body size at birth. Our data suggest that Tet1 loss leading to a partial reduction in 5hmC levels does not affect pluripotency in ESCs and is compatible with embryonic and postnatal development.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development.Dev Cell. 2013 Feb 11;24(3):310-23. doi: 10.1016/j.devcel.2012.12.015. Epub 2013 Jan 24. Dev Cell. 2013. PMID: 23352810 Free PMC article.
-
Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation.Nature. 2011 May 19;473(7347):398-402. doi: 10.1038/nature10008. Epub 2011 Apr 3. Nature. 2011. PMID: 21460836
-
Tet1 and 5-hydroxymethylation: a genome-wide view in mouse embryonic stem cells.Cell Cycle. 2011 Aug 1;10(15):2428-36. doi: 10.4161/cc.10.15.16930. Epub 2011 Aug 1. Cell Cycle. 2011. PMID: 21750410 Free PMC article.
-
Tet family proteins and 5-hydroxymethylcytosine in development and disease.Development. 2012 Jun;139(11):1895-902. doi: 10.1242/dev.070771. Development. 2012. PMID: 22569552 Free PMC article. Review.
-
Ten eleven translocation enzymes and 5-hydroxymethylation in mammalian development and cancer.Adv Exp Med Biol. 2013;754:57-79. doi: 10.1007/978-1-4419-9967-2_3. Adv Exp Med Biol. 2013. PMID: 22956496 Review.
Cited by
-
NANOG-dependent function of TET1 and TET2 in establishment of pluripotency.Nature. 2013 Mar 21;495(7441):370-4. doi: 10.1038/nature11925. Epub 2013 Feb 10. Nature. 2013. PMID: 23395962 Free PMC article.
-
Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond.Epigenetics Chromatin. 2015 Jul 21;8:24. doi: 10.1186/s13072-015-0016-6. eCollection 2015. Epigenetics Chromatin. 2015. PMID: 26195987 Free PMC article. Review.
-
Interrogating genomic and epigenomic data to understand prostate cancer.Biochim Biophys Acta. 2012 Apr;1825(2):186-96. doi: 10.1016/j.bbcan.2011.12.003. Epub 2012 Jan 3. Biochim Biophys Acta. 2012. PMID: 22240201 Free PMC article. Review.
-
The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells.Mol Cell. 2012 Dec 28;48(6):849-62. doi: 10.1016/j.molcel.2012.11.001. Epub 2012 Dec 6. Mol Cell. 2012. PMID: 23219530 Free PMC article.
-
The balance between NANOG and SOX17 mediated by TET proteins regulates specification of human primordial germ cell fate.Cell Biosci. 2022 Nov 4;12(1):181. doi: 10.1186/s13578-022-00917-0. Cell Biosci. 2022. PMID: 36333732 Free PMC article.
References
-
- Dawlaty MM, van Deursen JM. Gene targeting methods for studying nuclear transport factors in mice. Methods. 2006;39:370–378. - PubMed
-
- Dean W, Santos F, Reik W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin Cell Dev Biol. 2003;14:93–100. - PubMed
-
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 CA087869-10/CA/NCI NIH HHS/United States
- 5-R01-HDO45022/PHS HHS/United States
- R01 HD045022-06/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 5-R37-CA084198/CA/NCI NIH HHS/United States
- R37 HD045022/HD/NICHD NIH HHS/United States
- 5-R01-CA087869/CA/NCI NIH HHS/United States
- R01 CA087869/CA/NCI NIH HHS/United States
- R01 CA084198-13/CA/NCI NIH HHS/United States
- R37 CA084198/CA/NCI NIH HHS/United States
- R01 HD045022/HD/NICHD NIH HHS/United States
- R01 CA084198/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

