DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling
- PMID: 21791617
- PMCID: PMC3165724
- DOI: 10.1128/MCB.01368-10
DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling
Abstract
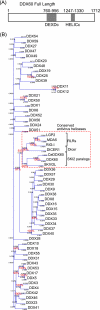
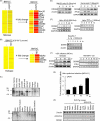
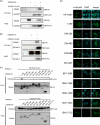
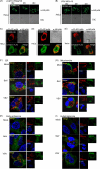
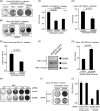
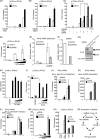
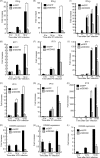
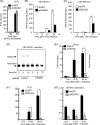
The cytoplasmic viral RNA sensors RIG-I and MDA5 are important for the production of type I interferon and other inflammatory cytokines. DDX60 is an uncharacterized DEXD/H box RNA helicase similar to Saccharomyces cerevisiae Ski2, a cofactor of RNA exosome, which is a protein complex required for the integrity of cytoplasmic RNA. Expression of DDX60 increases after viral infection, and the protein localizes at the cytoplasmic region. After viral infection, the DDX60 protein binds to endogenous RIG-I protein. The protein also binds to MDA5 and LGP2 but not to the downstream factors IPS-1 and IκB kinase ε (IKK-ε). Knockdown analysis shows that DDX60 is required for RIG-I- or MDA5-dependent type I interferon and interferon-inducible gene expression in response to viral infection. However, DDX60 is dispensable for TLR3-mediated signaling. Purified DDX60 helicase domains possess the activity to bind to viral RNA and DNA. Expression of DDX60 promotes the binding of RIG-I to double-stranded RNA. Taken together, our analyses indicate that DDX60 is a novel antiviral helicase promoting RIG-I-like receptor-mediated signaling.
Figures




Similar articles
-
DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential.Eur J Immunol. 2010 Apr;40(4):940-8. doi: 10.1002/eji.200940203. Eur J Immunol. 2010. PMID: 20127681
-
DDX60 Is Involved in RIG-I-Dependent and Independent Antiviral Responses, and Its Function Is Attenuated by Virus-Induced EGFR Activation.Cell Rep. 2015 May 26;11(8):1193-207. doi: 10.1016/j.celrep.2015.04.047. Epub 2015 May 14. Cell Rep. 2015. PMID: 25981042
-
LGP2 binds to PACT to regulate RIG-I- and MDA5-mediated antiviral responses.Sci Signal. 2019 Oct 1;12(601):eaar3993. doi: 10.1126/scisignal.aar3993. Sci Signal. 2019. PMID: 31575732
-
Retinoic acid-inducible gene-I-like receptors.J Interferon Cytokine Res. 2011 Jan;31(1):27-31. doi: 10.1089/jir.2010.0057. Epub 2010 Oct 15. J Interferon Cytokine Res. 2011. PMID: 20950133 Review.
-
Proofreading mechanisms of the innate immune receptor RIG-I: distinguishing self and viral RNA.Biochem Soc Trans. 2024 Jun 26;52(3):1131-1148. doi: 10.1042/BST20230724. Biochem Soc Trans. 2024. PMID: 38884803 Free PMC article. Review.
Cited by
-
Transcriptome Analysis of Duck and Chicken Brains Infected with Aquatic Bird Bornavirus-1 (ABBV-1).Viruses. 2022 Oct 8;14(10):2211. doi: 10.3390/v14102211. Viruses. 2022. PMID: 36298766 Free PMC article.
-
Possible involvement of DExD/H box helicase 60 in synovial inflammation of rheumatoid arthritis: role of toll-like receptor 3 signaling.Mol Biol Rep. 2024 Jan 18;51(1):131. doi: 10.1007/s11033-023-09063-3. Mol Biol Rep. 2024. PMID: 38236450
-
HSV-1-induced chemokine expression via IFI16-dependent and IFI16-independent pathways in human monocyte-derived macrophages.Herpesviridae. 2012 Oct 14;3(1):6. doi: 10.1186/2042-4280-3-6. Herpesviridae. 2012. PMID: 23062757 Free PMC article.
-
Interferon alpha inducible protein 6 is a negative regulator of innate immune responses by modulating RIG-I activation.Front Immunol. 2023 Jan 30;14:1105309. doi: 10.3389/fimmu.2023.1105309. eCollection 2023. Front Immunol. 2023. PMID: 36793726 Free PMC article.
-
Zika virus disrupts gene expression in human myoblasts and myotubes: Relationship with susceptibility to infection.PLoS Negl Trop Dis. 2022 Feb 16;16(2):e0010166. doi: 10.1371/journal.pntd.0010166. eCollection 2022 Feb. PLoS Negl Trop Dis. 2022. PMID: 35171909 Free PMC article.
References
-
- Brouwer R., et al. 2001. Three novel components of the human exosome. J. Biol. Chem. 276: 6177–6184 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
