Microarrays in the 2010s: the contribution of microarray-based gene expression profiling to breast cancer classification, prognostication and prediction
- PMID: 21787441
- PMCID: PMC3218943
- DOI: 10.1186/bcr2890
Microarrays in the 2010s: the contribution of microarray-based gene expression profiling to breast cancer classification, prognostication and prediction
Abstract
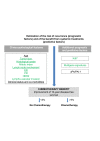
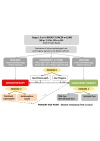
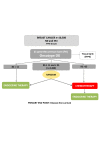
Breast cancer comprises a collection of diseases with distinctive clinical, histopathological, and molecular features. Importantly, tumors with similar histological features may display disparate clinical behaviors. Gene expression profiling using microarray technologies has improved our understanding of breast cancer biology and has led to the development of a breast cancer molecular taxonomy and of multigene 'signatures' to predict outcome and response to systemic therapies. The use of these prognostic and predictive signatures in routine clinical decision-making remains controversial. Here, we review the clinical relevance of microarray-based profiling of breast cancer and discuss its impact on patient management.
Figures
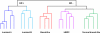
Similar articles
-
The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade.J Pathol. 2010 Jan;220(2):263-80. doi: 10.1002/path.2648. J Pathol. 2010. PMID: 19927298 Review.
-
Microarray-based gene expression profiling as a clinical tool for breast cancer management: are we there yet?Int J Surg Pathol. 2009 Aug;17(4):285-302. doi: 10.1177/1066896908328577. Epub 2008 Dec 22. Int J Surg Pathol. 2009. PMID: 19103611 Review.
-
Microarray technology and its effect on breast cancer (re)classification and prediction of outcome.Breast Cancer Res. 2003;5(6):303-4. doi: 10.1186/bcr732. Epub 2003 Oct 9. Breast Cancer Res. 2003. PMID: 14580246 Free PMC article. No abstract available.
-
Discerning Clinical Responses in Breast Cancer Based On Molecular Signatures.Am J Pathol. 2017 Oct;187(10):2199-2207. doi: 10.1016/j.ajpath.2017.08.002. Epub 2017 Aug 16. Am J Pathol. 2017. PMID: 28822803 Review.
-
Histological types of breast cancer: how special are they?Mol Oncol. 2010 Jun;4(3):192-208. doi: 10.1016/j.molonc.2010.04.004. Epub 2010 Apr 18. Mol Oncol. 2010. PMID: 20452298 Free PMC article. Review.
Cited by
-
Are breast cancer molecular classes predictive of survival in patients with long follow-up?Dis Markers. 2013;35(6):595-605. doi: 10.1155/2013/347073. Epub 2013 Oct 30. Dis Markers. 2013. PMID: 24288429 Free PMC article.
-
MVIAeval: a web tool for comprehensively evaluating the performance of a new missing value imputation algorithm.BMC Bioinformatics. 2017 Jan 13;18(1):31. doi: 10.1186/s12859-016-1429-3. BMC Bioinformatics. 2017. PMID: 28086746 Free PMC article.
-
Targeting Intracellular Calcium Signaling ([Ca2+]i) to Overcome Acquired Multidrug Resistance of Cancer Cells: A Mini-Overview.Cancers (Basel). 2017 May 9;9(5):48. doi: 10.3390/cancers9050048. Cancers (Basel). 2017. PMID: 28486397 Free PMC article. Review.
-
Screening-relevant age threshold of 70 years and older is a stronger determinant for the choice of adjuvant treatment in breast cancer patients than tumor biology.Breast Cancer Res Treat. 2017 May;163(1):119-130. doi: 10.1007/s10549-017-4151-6. Epub 2017 Feb 15. Breast Cancer Res Treat. 2017. PMID: 28205042 Free PMC article.
-
Genomic instability at the 13q31 locus and somatic mtDNA mutation in the D-loop site correlate with tumor aggressiveness in sporadic Brazilian breast cancer cases.Clinics (Sao Paulo). 2012 Oct;67(10):1181-90. doi: 10.6061/clinics/2012(10)10. Clinics (Sao Paulo). 2012. PMID: 23070345 Free PMC article.
References
-
- Reis-Filho JS, Weigelt B, Fumagalli D, Sotiriou C. Molecular profiling: moving away from tumor philately. Sci Transl Med. 2010;2:47ps43. - PubMed
-
- Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220:263–280. - PubMed
-
- Adjuvant! Online homepage. https://www.adjuvantonline.com
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

