Proteomic profiling of S-acylated macrophage proteins identifies a role for palmitoylation in mitochondrial targeting of phospholipid scramblase 3
- PMID: 21785166
- PMCID: PMC3205854
- DOI: 10.1074/mcp.M110.006007
Proteomic profiling of S-acylated macrophage proteins identifies a role for palmitoylation in mitochondrial targeting of phospholipid scramblase 3
Abstract
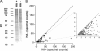
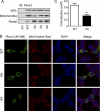
S-Palmitoylation, the reversible post-translational acylation of specific cysteine residues with the fatty acid palmitate, promotes the membrane tethering and subcellular localization of proteins in several biological pathways. Although inhibiting palmitoylation holds promise as a means for manipulating protein targeting, advances in the field have been hampered by limited understanding of palmitoylation enzymology and consensus motifs. In order to define the complement of S-acylated proteins in the macrophage, we treated RAW 264.7 macrophage membranes with hydroxylamine to cleave acyl thioesters, followed by biotinylation of newly exposed sulfhydryls and streptavidin-agarose affinity chromatography. Among proteins identified by LC-MS/MS, S-acylation status was established by spectral counting to assess enrichment under hydroxylamine versus mock treatment conditions. Of 1183 proteins identified in four independent experiments, 80 proteins were significant for S-acylation at false discovery rate = 0.05, and 101 significant at false discovery rate = 0.10. Candidate S-acylproteins were identified from several functional categories, including membrane trafficking, signaling, transporters, and receptors. Among these were 29 proteins previously biochemically confirmed as palmitoylated, 45 previously reported as putative S-acylproteins in proteomic screens, 24 not previously associated with palmitoylation, and three presumed false-positives. Nearly half of the candidates were previously identified by us in macrophage detergent-resistant membranes, suggesting that palmitoylation promotes lipid raft-localization of proteins in the macrophage. Among the candidate novel S-acylproteins was phospholipid scramblase 3 (Plscr3), a protein that regulates apoptosis through remodeling the mitochondrial membrane. Palmitoylation of Plscr3 was confirmed through (3)H-palmitate labeling. Moreover, site-directed mutagenesis of a cluster of five cysteines (Cys159-161-163-164-166) abolished palmitoylation, caused Plscr3 mislocalization from mitochondrion to nucleus, and reduced macrophage apoptosis in response to etoposide, together suggesting a role for palmitoylation at this site for mitochondrial targeting and pro-apoptotic function of Plscr3. Taken together, we propose that manipulation of protein palmitoylation carries great potential for intervention in macrophage biology via reprogramming of protein localization.
Figures



Similar articles
-
Selective Enrichment and Direct Analysis of Protein S-Palmitoylation Sites.J Proteome Res. 2018 May 4;17(5):1907-1922. doi: 10.1021/acs.jproteome.8b00002. Epub 2018 Apr 6. J Proteome Res. 2018. PMID: 29575903 Free PMC article.
-
Chemical Proteomic Analysis of S-Fatty Acylated Proteins and Their Modification Sites.Methods Mol Biol. 2019;2009:45-57. doi: 10.1007/978-1-4939-9532-5_4. Methods Mol Biol. 2019. PMID: 31152394
-
Insulin-regulated protein palmitoylation impacts endothelial cell function.Arterioscler Thromb Vasc Biol. 2014 Feb;34(2):346-54. doi: 10.1161/ATVBAHA.113.302848. Epub 2013 Dec 19. Arterioscler Thromb Vasc Biol. 2014. PMID: 24357059 Free PMC article.
-
Protein S-palmitoylation in cellular differentiation.Biochem Soc Trans. 2017 Feb 8;45(1):275-285. doi: 10.1042/BST20160236. Biochem Soc Trans. 2017. PMID: 28202682 Free PMC article. Review.
-
Methodology for Detecting Protein Palmitoylation.Adv Exp Med Biol. 2020;1248:425-430. doi: 10.1007/978-981-15-3266-5_17. Adv Exp Med Biol. 2020. PMID: 32185720 Review.
Cited by
-
Examining the Underappreciated Role of S-Acylated Proteins as Critical Regulators of Phagocytosis and Phagosome Maturation in Macrophages.Front Immunol. 2021 Apr 1;12:659533. doi: 10.3389/fimmu.2021.659533. eCollection 2021. Front Immunol. 2021. PMID: 33868308 Free PMC article. Review.
-
Stress-induced Changes in the S-palmitoylation and S-nitrosylation of Synaptic Proteins.Mol Cell Proteomics. 2019 Oct;18(10):1916-1938. doi: 10.1074/mcp.RA119.001581. Epub 2019 Jul 16. Mol Cell Proteomics. 2019. PMID: 31311849 Free PMC article.
-
p53 Integrates host defense and cell fate during bacterial pneumonia.J Exp Med. 2013 May 6;210(5):891-904. doi: 10.1084/jem.20121674. Epub 2013 Apr 29. J Exp Med. 2013. PMID: 23630228 Free PMC article.
-
Identification and characterisation of a phospholipid scramblase in the malaria parasite Plasmodium falciparum.Mol Biochem Parasitol. 2021 May;243:111374. doi: 10.1016/j.molbiopara.2021.111374. Epub 2021 May 8. Mol Biochem Parasitol. 2021. PMID: 33974939 Free PMC article.
-
Palmitoylated calnexin is a key component of the ribosome-translocon complex.EMBO J. 2012 Apr 4;31(7):1823-35. doi: 10.1038/emboj.2012.15. Epub 2012 Feb 7. EMBO J. 2012. PMID: 22314232 Free PMC article.
References
-
- Iwanaga T., Tsutsumi R., Noritake J., Fukata Y., Fukata M. (2009) Dynamic protein palmitoylation in cellular signaling. Prog. Lipid Res. 48, 117–127 - PubMed
-
- Resh M. D. (2006) Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE. 2006, re14. - PubMed
-
- Wedegaertner P. B., Bourne H. R. (1994) Activation and depalmitoylation of Gs alpha. Cell 77, 1063–1070 - PubMed
-
- El-Husseini Ael-D., Schnell E., Dakoji S., Sweeney N., Zhou Q., Prange O., Gauthier-Campbell C., Aguilera-Moreno A., Nicoll R. A., Bredt D. S. (2002) Synaptic strength regulated by palmitate cycling on PSD-95. Cell 108, 849–863 - PubMed
-
- Kostiuk M. A., Keller B. O., Berthiaume L. G. (2009) Non-radioactive detection of palmitoylated mitochondrial proteins using an azido-palmitate analogue. Methods Enzymol. 457, 149–165 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

