Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony
- PMID: 21779173
- PMCID: PMC3136464
- DOI: 10.1371/journal.ppat.1002155
Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony
Erratum in
- PLoS Pathog. 2011 Aug;7(8), doi:10.1371/annotation/59703f7f-9506-49d1-b339-09ee31510e89
- PLoS Pathog. 2011 Aug;7(8); doi:10.1371/annotation/c9a506d7-e8ba-4aaf-ab04-8ed0b383f5d9. Tarara, Ross P [added]; Canfield, Don R [added]
Abstract
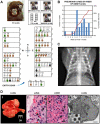
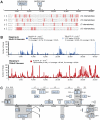
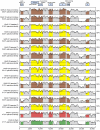
Adenoviruses are DNA viruses that naturally infect many vertebrates, including humans and monkeys, and cause a wide range of clinical illnesses in humans. Infection from individual strains has conventionally been thought to be species-specific. Here we applied the Virochip, a pan-viral microarray, to identify a novel adenovirus (TMAdV, titi monkey adenovirus) as the cause of a deadly outbreak in a closed colony of New World monkeys (titi monkeys; Callicebus cupreus) at the California National Primate Research Center (CNPRC). Among 65 titi monkeys housed in a building, 23 (34%) developed upper respiratory symptoms that progressed to fulminant pneumonia and hepatitis, and 19 of 23 monkeys, or 83% of those infected, died or were humanely euthanized. Whole-genome sequencing of TMAdV revealed that this adenovirus is a new species and highly divergent, sharing <57% pairwise nucleotide identity with other adenoviruses. Cultivation of TMAdV was successful in a human A549 lung adenocarcinoma cell line, but not in primary or established monkey kidney cells. At the onset of the outbreak, the researcher in closest contact with the monkeys developed an acute respiratory illness, with symptoms persisting for 4 weeks, and had a convalescent serum sample seropositive for TMAdV. A clinically ill family member, despite having no contact with the CNPRC, also tested positive, and screening of a set of 81 random adult blood donors from the Western United States detected TMAdV-specific neutralizing antibodies in 2 individuals (2/81, or 2.5%). These findings raise the possibility of zoonotic infection by TMAdV and human-to-human transmission of the virus in the population. Given the unusually high case fatality rate from the outbreak (83%), it is unlikely that titi monkeys are the native host species for TMAdV, and the natural reservoir of the virus is still unknown. The discovery of TMAdV, a novel adenovirus with the capacity to infect both monkeys and humans, suggests that adenoviruses should be monitored closely as potential causes of cross-species outbreaks.
Conflict of interest statement
The authors received a viral discovery award from Abbott Diagnostics (to CYC). The University of California, San Francisco (UCSF) has also filed a patent application related to TMAdV. This does not alter the authors' adherence to all PloS Pathogens policies on sharing data and materials.
Figures


Similar articles
-
Experimental cross-species infection of common marmosets by titi monkey adenovirus.PLoS One. 2013 Jul 24;8(7):e68558. doi: 10.1371/journal.pone.0068558. Print 2013. PLoS One. 2013. PMID: 23894316 Free PMC article.
-
Epidemiological and molecular characterization of a novel adenovirus of squirrel monkeys after fatal infection during immunosuppression.Microb Genom. 2020 Sep;6(9):mgen000395. doi: 10.1099/mgen.0.000395. Epub 2020 Jul 2. Microb Genom. 2020. PMID: 32614763 Free PMC article.
-
A novel adenovirus species associated with an acute respiratory outbreak in a baboon colony and evidence of coincident human infection.mBio. 2013 Apr 16;4(2):e00084. doi: 10.1128/mBio.00084-13. mBio. 2013. PMID: 23592261 Free PMC article.
-
[Mechanisms of viral emergence and interspecies transmission: the exemple of simian foamy viruses in Central Africa].Bull Acad Natl Med. 2013 Dec;197(9):1655-67; discussion 1667-8. doi: 10.1016/S0001-4079(19)31387-1. Bull Acad Natl Med. 2013. PMID: 26137812 Free PMC article. Review. French.
-
Adenoviruses in the immunocompromised host.Clin Microbiol Rev. 1992 Jul;5(3):262-74. doi: 10.1128/CMR.5.3.262. Clin Microbiol Rev. 1992. PMID: 1323383 Free PMC article. Review.
Cited by
-
Two novel adenoviruses found in Cave Myotis bats (Myotis velifer) in Oklahoma.Virus Genes. 2020 Feb;56(1):99-103. doi: 10.1007/s11262-019-01719-2. Epub 2019 Dec 3. Virus Genes. 2020. PMID: 31797220 Free PMC article.
-
Target-dependent enrichment of virions determines the reduction of high-throughput sequencing in virus discovery.PLoS One. 2015 Apr 8;10(4):e0122636. doi: 10.1371/journal.pone.0122636. eCollection 2015. PLoS One. 2015. PMID: 25853649 Free PMC article.
-
Evaluation of bat adenoviruses suggests co-evolution and host roosting behaviour as drivers for diversity.Microb Genom. 2021 Apr;7(4):000561. doi: 10.1099/mgen.0.000561. Microb Genom. 2021. PMID: 33871330 Free PMC article.
-
High Diversity and Novel Enteric Viruses in Fecal Viromes of Healthy Wild and Captive Thai Cynomolgus Macaques (Macaca fascicularis).Viruses. 2019 Oct 22;11(10):971. doi: 10.3390/v11100971. Viruses. 2019. PMID: 31652508 Free PMC article.
-
Community health and human-animal contacts on the edges of Bwindi Impenetrable National Park, Uganda.PLoS One. 2021 Nov 24;16(11):e0254467. doi: 10.1371/journal.pone.0254467. eCollection 2021. PLoS One. 2021. PMID: 34818325 Free PMC article.
References
-
- Harrach B, Benkõ M, Both G, Brown M, Davison A, et al. Family Adenoviridae. In: King A, Carstens E, Adams M, Lefkowitz E, editors. Virus Taxonomy: 9th Report of the International Committee on Taxonomy of Viruses. New York: Elsevier; 2011.
-
- Fox JP, Hall CE, Cooney MK. The Seattle Virus Watch. VII. Observations of adenovirus infections. Am J Epidemiol. 1977;105:362–386. - PubMed
-
- Lewis PF, Schmidt MA, Lu X, Erdman DD, Campbell M, et al. A community-based outbreak of severe respiratory illness caused by human adenovirus serotype 14. J Infect Dis. 2009;199:1427–1434. - PubMed
-
- Ruuskanen O, Meurman O, Akusjarvi G. Adenoviruses. In: Richman DD, Whitley RJ, Hayden FG, editors. Clinical Virology. New York: Churchill Livingstone; 1997. 1355 xvi.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

