Reversible inhibition of murine cytomegalovirus replication by gamma interferon (IFN-γ) in primary macrophages involves a primed type I IFN-signaling subnetwork for full establishment of an immediate-early antiviral state
- PMID: 21775459
- PMCID: PMC3196417
- DOI: 10.1128/JVI.00373-11
Reversible inhibition of murine cytomegalovirus replication by gamma interferon (IFN-γ) in primary macrophages involves a primed type I IFN-signaling subnetwork for full establishment of an immediate-early antiviral state
Abstract
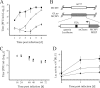
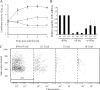
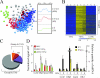
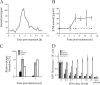
Activated macrophages play a central role in controlling inflammatory responses to infection and are tightly regulated to rapidly mount responses to infectious challenge. Type I interferon (alpha/beta interferon [IFN-α/β]) and type II interferon (IFN-γ) play a crucial role in activating macrophages and subsequently restricting viral infections. Both types of IFNs signal through related but distinct signaling pathways, inducing a vast number of interferon-stimulated genes that are overlapping but distinguishable. The exact mechanism by which IFNs, particularly IFN-γ, inhibit DNA viruses such as cytomegalovirus (CMV) is still not fully understood. Here, we investigate the antiviral state developed in macrophages upon reversible inhibition of murine CMV by IFN-γ. On the basis of molecular profiling of the reversible inhibition, we identify a significant contribution of a restricted type I IFN subnetwork linked with IFN-γ activation. Genetic knockout of the type I-signaling pathway, in the context of IFN-γ stimulation, revealed an essential requirement for a primed type I-signaling process in developing a full refractory state in macrophages. A minimal transient induction of IFN-β upon macrophage activation with IFN-γ is also detectable. In dose and kinetic viral replication inhibition experiments with IFN-γ, the establishment of an antiviral effect is demonstrated to occur within the first hours of infection. We show that the inhibitory mechanisms at these very early times involve a blockade of the viral major immediate-early promoter activity. Altogether our results show that a primed type I IFN subnetwork contributes to an immediate-early antiviral state induced by type II IFN activation of macrophages, with a potential further amplification loop contributed by transient induction of IFN-β.
Figures


Similar articles
-
Neuronal Ablation of Alpha/Beta Interferon (IFN-α/β) Signaling Exacerbates Central Nervous System Viral Dissemination and Impairs IFN-γ Responsiveness in Microglia/Macrophages.J Virol. 2020 Sep 29;94(20):e00422-20. doi: 10.1128/JVI.00422-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32796063 Free PMC article.
-
Murine cytomegalovirus infection inhibits IFN gamma-induced MHC class II expression on macrophages: the role of type I interferon.Virology. 1998 Feb 15;241(2):331-44. doi: 10.1006/viro.1997.8969. Virology. 1998. PMID: 9499808
-
Alpha/Beta Interferon (IFN-α/β) Signaling in Astrocytes Mediates Protection against Viral Encephalomyelitis and Regulates IFN-γ-Dependent Responses.J Virol. 2018 Apr 27;92(10):e01901-17. doi: 10.1128/JVI.01901-17. Print 2018 May 15. J Virol. 2018. PMID: 29491163 Free PMC article.
-
Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis.PLoS Biol. 2011 Mar;9(3):e1000598. doi: 10.1371/journal.pbio.1000598. Epub 2011 Mar 8. PLoS Biol. 2011. PMID: 21408089 Free PMC article.
-
Interferon gamma regulates acute and latent murine cytomegalovirus infection and chronic disease of the great vessels.J Exp Med. 1998 Aug 3;188(3):577-88. doi: 10.1084/jem.188.3.577. J Exp Med. 1998. PMID: 9687534 Free PMC article.
Cited by
-
Ablation of the regulatory IE1 protein of murine cytomegalovirus alters in vivo pro-inflammatory TNF-alpha production during acute infection.PLoS Pathog. 2012;8(8):e1002901. doi: 10.1371/journal.ppat.1002901. Epub 2012 Aug 30. PLoS Pathog. 2012. PMID: 22952450 Free PMC article.
-
Cytomegalovirus memory inflation and immune protection.Med Microbiol Immunol. 2019 Aug;208(3-4):339-347. doi: 10.1007/s00430-019-00607-8. Epub 2019 Apr 10. Med Microbiol Immunol. 2019. PMID: 30972476 Review.
-
STAT2-Dependent Immune Responses Ensure Host Survival despite the Presence of a Potent Viral Antagonist.J Virol. 2018 Jun 29;92(14):e00296-18. doi: 10.1128/JVI.00296-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29743368 Free PMC article.
-
Cytomegaloviruses and Macrophages-Friends and Foes From Early on?Front Immunol. 2020 May 12;11:793. doi: 10.3389/fimmu.2020.00793. eCollection 2020. Front Immunol. 2020. PMID: 32477336 Free PMC article. Review.
-
The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response.Immunity. 2013 Jan 24;38(1):106-18. doi: 10.1016/j.immuni.2012.11.004. Epub 2012 Dec 27. Immunity. 2013. PMID: 23273843 Free PMC article.
References
-
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

