Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress
- PMID: 21768361
- PMCID: PMC3150939
- DOI: 10.1073/pnas.1109796108
Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress
Abstract
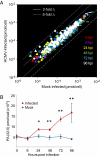
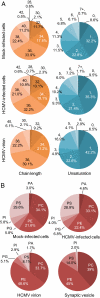
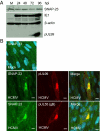
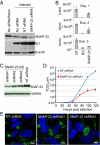
Human cytomegalovirus induces and requires fatty acid synthesis. This suggests an essential role for lipidome remodeling in viral replication. We used mass spectrometry to quantify glycerophospholipids in mock-infected and virus-infected fibroblasts, as well as in virions. Although the lipid composition of mock-infected and virus-infected fibroblasts was similar, virions were markedly different. The virion envelope contained twofold more phosphatidylethanolamines and threefold less phosphatidylserines than the host cell. This indicates that the virus buds from a membrane with a different lipid composition from the host cell as a whole. Compared with published datasets, the virion envelope showed the greatest similarity to the synaptic vesicle lipidome. Synaptosome-associated protein of 25 kDa (SNAP-25) is a component of the complex that mediates exocytosis of synaptic vesicles in neurons; and its homolog, SNAP-23, functions in exocytosis in many other cell types. Infection induced the relocation of SNAP-23 to the cytoplasmic viral assembly zone, and knockdown of SNAP-23 inhibited the production of virus. We propose that cytomegalovirus capsids acquire their envelope by budding into vesicles with a lipid composition similar to that of synaptic vesicles, which subsequently fuse with the plasma membrane to release virions from the cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Identification of soluble N-ethylmaleimide-sensitive factor attachment protein receptor exocytotic machinery in human plasma cells: SNAP-23 is essential for antibody secretion.J Immunol. 2005 Nov 15;175(10):6686-93. doi: 10.4049/jimmunol.175.10.6686. J Immunol. 2005. PMID: 16272324
-
Roles of Cellular NSF Protein in Entry and Nuclear Egress of Budded Virions of Autographa californica Multiple Nucleopolyhedrovirus.J Virol. 2017 Sep 27;91(20):e01111-17. doi: 10.1128/JVI.01111-17. Print 2017 Oct 15. J Virol. 2017. PMID: 28747507 Free PMC article.
-
SNAP-23 and VAMP-3 contribute to the release of IL-6 and TNFα from a human synovial sarcoma cell line.FEBS J. 2014 Feb;281(3):750-65. doi: 10.1111/febs.12620. Epub 2013 Dec 13. FEBS J. 2014. PMID: 24373201
-
The assembly of lipid droplets and its relation to cellular insulin sensitivity.Biochem Soc Trans. 2009 Oct;37(Pt 5):981-5. doi: 10.1042/BST0370981. Biochem Soc Trans. 2009. PMID: 19754436 Review.
-
SNARE complex regulation by phosphorylation.Cell Biochem Biophys. 2006;45(1):111-23. doi: 10.1385/CBB:45:1:111. Cell Biochem Biophys. 2006. PMID: 16679567 Review.
Cited by
-
Lipidomics: when apocrypha becomes canonical.Curr Opin Chem Biol. 2012 Apr;16(1-2):221-6. doi: 10.1016/j.cbpa.2012.02.003. Epub 2012 Feb 28. Curr Opin Chem Biol. 2012. PMID: 22381642 Free PMC article. Review.
-
Assembly-hub function of ER-localized SNARE proteins in biogenesis of tombusvirus replication compartment.PLoS Pathog. 2018 May 10;14(5):e1007028. doi: 10.1371/journal.ppat.1007028. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29746582 Free PMC article.
-
Manipulation of host pathways by human cytomegalovirus: insights from genome-wide studies.Semin Immunopathol. 2014 Nov;36(6):651-8. doi: 10.1007/s00281-014-0443-7. Epub 2014 Sep 27. Semin Immunopathol. 2014. PMID: 25260940 Review.
-
Metabolomics in drug target discovery.Cold Spring Harb Symp Quant Biol. 2011;76:235-46. doi: 10.1101/sqb.2011.76.010694. Epub 2011 Nov 23. Cold Spring Harb Symp Quant Biol. 2011. PMID: 22114327 Free PMC article. Review.
-
Labyrinthopeptins Exert Broad-Spectrum Antiviral Activity through Lipid-Binding-Mediated Virolysis.J Virol. 2020 Jan 6;94(2):e01471-19. doi: 10.1128/JVI.01471-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31666384 Free PMC article.
References
-
- Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

