Mechanism of 150-cavity formation in influenza neuraminidase
- PMID: 21750542
- PMCID: PMC3144582
- DOI: 10.1038/ncomms1390
Mechanism of 150-cavity formation in influenza neuraminidase
Abstract
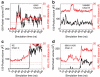
The recently discovered 150-cavity in the active site of group-1 influenza A neuraminidase (NA) proteins provides a target for rational structure-based drug development to counter the increasing frequency of antiviral resistance in influenza. Surprisingly, the 2009 H1N1 pandemic virus (09N1) neuraminidase was crystalized without the 150-cavity characteristic of group-1 NAs. Here we demonstrate, through a total sum of 1.6 μs of biophysical simulations, that 09N1 NA exists in solution preferentially with an open 150-cavity. Comparison with simulations using avian N1, human N2 and 09N1 with a I149V mutation and an extensive bioinformatics analysis suggests that the conservation of a key salt bridge is crucial in the stabilization of the 150-cavity across both subtypes. This result provides an atomic-level structural understanding of the recent finding that antiviral compounds designed to take advantage of contacts in the 150-cavity can inactivate both 2009 H1N1 pandemic and avian H5N1 viruses.
Figures

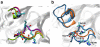
Similar articles
-
Antiviral susceptibility of avian and swine influenza virus of the N1 neuraminidase subtype.J Virol. 2010 Oct;84(19):9800-9. doi: 10.1128/JVI.00296-10. Epub 2010 Jul 21. J Virol. 2010. PMID: 20660186 Free PMC article.
-
[Virological impact of stalk region of neuraminidase in influenza A/Anhui/1/05 (H5N1) and A/Ohio/07/2009 (H1N1) viruses].Bing Du Xue Bao. 2014 May;30(3):238-45. Bing Du Xue Bao. 2014. PMID: 25118377 Chinese.
-
Characterization of neuraminidases from the highly pathogenic avian H5N1 and 2009 pandemic H1N1 influenza A viruses.PLoS One. 2010 Dec 29;5(12):e15825. doi: 10.1371/journal.pone.0015825. PLoS One. 2010. PMID: 21209916 Free PMC article.
-
Computational studies of influenza A virus at three important targets: hemagglutinin, neuraminidase and M2 protein.Curr Pharm Des. 2011;17(17):1720-39. doi: 10.2174/138161211796355083. Curr Pharm Des. 2011. PMID: 21619529 Review.
-
Recent advances in neuraminidase inhibitor development as anti-influenza drugs.ChemMedChem. 2012 Sep;7(9):1527-36. doi: 10.1002/cmdc.201200155. Epub 2012 Jul 16. ChemMedChem. 2012. PMID: 22807317 Review.
Cited by
-
A virtual screening approach for identifying plants with anti H5N1 neuraminidase activity.J Chem Inf Model. 2015 Feb 23;55(2):308-16. doi: 10.1021/ci500405g. Epub 2015 Jan 29. J Chem Inf Model. 2015. PMID: 25555059 Free PMC article.
-
Improving the Efficiency of Free Energy Calculations in the Amber Molecular Dynamics Package.J Chem Theory Comput. 2013 Sep 10;9(9):10.1021/ct400340s. doi: 10.1021/ct400340s. J Chem Theory Comput. 2013. PMID: 24185531 Free PMC article.
-
Influenza neuraminidase.Influenza Other Respir Viruses. 2012 Jul;6(4):245-56. doi: 10.1111/j.1750-2659.2011.00304.x. Epub 2011 Nov 16. Influenza Other Respir Viruses. 2012. PMID: 22085243 Free PMC article. Review.
-
Microsecond Molecular Dynamics Simulations of Influenza Neuraminidase Suggest a Mechanism for the Increased Virulence of Stalk-Deletion Mutants.J Phys Chem B. 2016 Aug 25;120(33):8590-9. doi: 10.1021/acs.jpcb.6b02655. Epub 2016 May 12. J Phys Chem B. 2016. PMID: 27141956 Free PMC article.
-
Exploring the chemical space of influenza neuraminidase inhibitors.PeerJ. 2016 Apr 19;4:e1958. doi: 10.7717/peerj.1958. eCollection 2016. PeerJ. 2016. PMID: 27114890 Free PMC article.
References
-
- Russell R. J. et al.. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443, 45–49 (2006). - PubMed
-
- von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 6, 967–974 (2007). - PubMed
-
- Dharan N. J. et al.. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 301, 1034–1041 (2009). - PubMed
-
- Li Q. et al.. The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat. Struct. Mol. Biol. 17, 1266–1268 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

