Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth
- PMID: 21746931
- PMCID: PMC3169132
- DOI: 10.1073/pnas.1108892108
Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth
Abstract
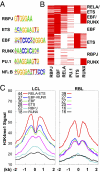
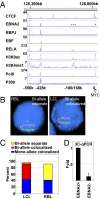
Epstein-Barr virus nuclear antigen 2 (EBNA2) regulation of transcription through the cell transcription factor RBPJ is essential for resting B-lymphocyte (RBL) conversion to immortal lymphoblast cell lines (LCLs). ChIP-seq of EBNA2 and RBPJ sites in LCL DNA found EBNA2 at 5,151 and RBPJ at 10,529 sites. EBNA2 sites were enriched for RBPJ (78%), early B-cell factor (EBF, 39%), RUNX (43%), ETS (39%), NFκB (22%), and PU.1 (22%) motifs. These motif associations were confirmed by LCL RBPJ ChIP-seq finding 72% RBPJ occupancy and Encyclopedia Of DNA Elements LCL ChIP-seq finding EBF, NFκB RELA, and PU.1 at 54%, 31%, and 17% of EBNA2 sites. EBNA2 and RBPJ were predominantly at intergene and intron sites and only 14% at promoter sites. K-means clustering of EBNA2 site transcription factors identified RELA-ETS, EBF-RUNX, EBF, ETS, RBPJ, and repressive RUNX clusters, which ranked from highest to lowest in H3K4me1 signals and nucleosome depletion, indicative of active chromatin. Surprisingly, although quantitatively less, the same genome sites in RBLs exhibited similar high-level H3K4me1 signals and nucleosome depletion. The EBV genome also had an LMP1 promoter EBF site, which proved critical for EBNA2 activation. LCL HiC data mapped intergenic EBNA2 sites to EBNA2 up-regulated genes. FISH and chromatin conformation capture linked EBNA2/RBPJ enhancers 428 kb 5' of MYC to MYC. These data indicate that EBNA2 evolved to target RBL H3K4me1 modified, nucleosome-depleted, nonpromoter sites to drive B-lymphocyte proliferation in primary human infection. The primed RBL program likely supports antigen-induced proliferation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Transcription factor RBPJ/CSL: a genome-wide look at transcriptional regulation.Proc Natl Acad Sci U S A. 2011 Sep 6;108(36):14715-6. doi: 10.1073/pnas.1110570108. Epub 2011 Aug 24. Proc Natl Acad Sci U S A. 2011. PMID: 21873209 Free PMC article. No abstract available.
Similar articles
-
Epstein-Barr Virus Nuclear Antigen 3 (EBNA3) Proteins Regulate EBNA2 Binding to Distinct RBPJ Genomic Sites.J Virol. 2015 Dec 30;90(6):2906-19. doi: 10.1128/JVI.02737-15. J Virol. 2015. PMID: 26719268 Free PMC article.
-
EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding.Proc Natl Acad Sci U S A. 2011 May 10;108(19):7808-13. doi: 10.1073/pnas.1104991108. Epub 2011 Apr 25. Proc Natl Acad Sci U S A. 2011. PMID: 21518914 Free PMC article.
-
Epstein-Barr virus nuclear antigen 3A partially coincides with EBNA3C genome-wide and is tethered to DNA through BATF complexes.Proc Natl Acad Sci U S A. 2015 Jan 13;112(2):554-9. doi: 10.1073/pnas.1422580112. Epub 2014 Dec 24. Proc Natl Acad Sci U S A. 2015. PMID: 25540416 Free PMC article.
-
Roles of RUNX in B Cell Immortalisation.Adv Exp Med Biol. 2017;962:283-298. doi: 10.1007/978-981-10-3233-2_18. Adv Exp Med Biol. 2017. PMID: 28299664 Review.
-
EBNA2 and Its Coactivator EBNA-LP.Curr Top Microbiol Immunol. 2015;391:35-59. doi: 10.1007/978-3-319-22834-1_2. Curr Top Microbiol Immunol. 2015. PMID: 26428371 Review.
Cited by
-
Regulation of B cell receptor signalling by Epstein-Barr virus nuclear antigens.Biochem J. 2022 Dec 9;479(23):2395-2417. doi: 10.1042/BCJ20220417. Biochem J. 2022. PMID: 36383217 Free PMC article.
-
Viral Proteins with PxxP and PY Motifs May Play a Role in Multiple Sclerosis.Viruses. 2022 Jan 28;14(2):281. doi: 10.3390/v14020281. Viruses. 2022. PMID: 35215874 Free PMC article. Review.
-
The Varied Roles of Notch in Cancer.Annu Rev Pathol. 2017 Jan 24;12:245-275. doi: 10.1146/annurev-pathol-052016-100127. Epub 2016 Dec 5. Annu Rev Pathol. 2017. PMID: 27959635 Free PMC article. Review.
-
Epstein-Barr Virus Episome Physically Interacts with Active Regions of the Host Genome in Lymphoblastoid Cells.J Virol. 2020 Nov 23;94(24):e01390-20. doi: 10.1128/JVI.01390-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32999023 Free PMC article.
-
Identification of a recurrent transforming UBR5-ZNF423 fusion gene in EBV-associated nasopharyngeal carcinoma.J Pathol. 2013 Oct;231(2):158-67. doi: 10.1002/path.4240. J Pathol. 2013. PMID: 23878065 Free PMC article.
References
-
- Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet. 1964;1:702–703. - PubMed
-
- Johansson B, Klein G, Henle W, Henle G. Epstein-Barr virus (EBV)-associated antibody patterns in malignant lymphoma and leukemia. I. Hodgkin's disease. Int J Cancer. 1970;6:450–462. - PubMed
-
- Rickinson A, Kieff E. Epstein-Barr Virus. In: Howley P, Knipe D, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2655–2700.
-
- Henle W, Diehl V, Kohn G, Zur Hausen H, Henle G. Herpes-type virus and chromosome marker in normal leukocytes after growth with irradiated Burkitt cells. Science. 1967;157:1064–1065. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

