Tracking genome engineering outcome at individual DNA breakpoints
- PMID: 21743461
- PMCID: PMC3415300
- DOI: 10.1038/nmeth.1648
Tracking genome engineering outcome at individual DNA breakpoints
Abstract
Site-specific genome engineering technologies are increasingly important tools in the postgenomic era, where biotechnological objectives often require organisms with precisely modified genomes. Rare-cutting endonucleases, through their capacity to create a targeted DNA strand break, are one of the most promising of these technologies. However, realizing the full potential of nuclease-induced genome engineering requires a detailed understanding of the variables that influence resolution of nuclease-induced DNA breaks. Here we present a genome engineering reporter system, designated 'traffic light', that supports rapid flow-cytometric analysis of repair pathway choice at individual DNA breaks, quantitative tracking of nuclease expression and donor template delivery, and high-throughput screens for factors that bias the engineering outcome. We applied the traffic light system to evaluate the efficiency and outcome of nuclease-induced genome engineering in human cell lines and identified strategies to facilitate isolation of cells in which a desired engineering outcome has occurred.
Conflict of interest statement
Figures
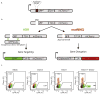
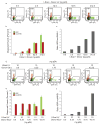
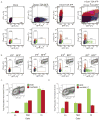

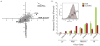
Comment in
-
Seeing the light: integrating genome engineering with double-strand break repair.Nat Methods. 2011 Jul 28;8(8):628-30. doi: 10.1038/nmeth.1656. Nat Methods. 2011. PMID: 21799496 No abstract available.
Similar articles
-
Seeing the light: integrating genome engineering with double-strand break repair.Nat Methods. 2011 Jul 28;8(8):628-30. doi: 10.1038/nmeth.1656. Nat Methods. 2011. PMID: 21799496 No abstract available.
-
DNA Break Repair in Plants and Its Application for Genome Engineering.Methods Mol Biol. 2019;1864:237-266. doi: 10.1007/978-1-4939-8778-8_17. Methods Mol Biol. 2019. PMID: 30415341
-
Novel fluorescent genome editing reporters for monitoring DNA repair pathway utilization at endonuclease-induced breaks.Nucleic Acids Res. 2014 Jan;42(1):e4. doi: 10.1093/nar/gkt872. Epub 2013 Oct 10. Nucleic Acids Res. 2014. PMID: 24121685 Free PMC article.
-
Genome engineering with TAL-effector nucleases and alternative modular nuclease technologies.Curr Gene Ther. 2013 Aug;13(4):291-303. doi: 10.2174/15665232113139990026. Curr Gene Ther. 2013. PMID: 23888878 Review.
-
Recent advances in targeted genome engineering in mammalian systems.Biotechnol J. 2012 Sep;7(9):1074-87. doi: 10.1002/biot.201200038. Epub 2012 Jul 10. Biotechnol J. 2012. PMID: 22777886 Review.
Cited by
-
TGF-β Inhibition Rescues Hematopoietic Stem Cell Defects and Bone Marrow Failure in Fanconi Anemia.Cell Stem Cell. 2016 May 5;18(5):668-81. doi: 10.1016/j.stem.2016.03.002. Epub 2016 Mar 24. Cell Stem Cell. 2016. PMID: 27053300 Free PMC article.
-
The COP9 signalosome is vital for timely repair of DNA double-strand breaks.Nucleic Acids Res. 2015 May 19;43(9):4517-30. doi: 10.1093/nar/gkv270. Epub 2015 Apr 8. Nucleic Acids Res. 2015. PMID: 25855810 Free PMC article.
-
Plasmid-based complementation of large deletions in Phaeodactylum tricornutum biosynthetic genes generated by Cas9 editing.Sci Rep. 2020 Aug 17;10(1):13879. doi: 10.1038/s41598-020-70769-6. Sci Rep. 2020. PMID: 32807825 Free PMC article.
-
pEVL: A Linear Plasmid for Generating mRNA IVT Templates With Extended Encoded Poly(A) Sequences.Mol Ther Nucleic Acids. 2016 Apr 19;5(4):e306. doi: 10.1038/mtna.2016.21. Mol Ther Nucleic Acids. 2016. PMID: 27093168 Free PMC article.
-
Short Double-Stranded DNA (≤40-bp) Affects Repair Pathway Choice.Int J Mol Sci. 2023 Jul 23;24(14):11836. doi: 10.3390/ijms241411836. Int J Mol Sci. 2023. PMID: 37511594 Free PMC article.
References
-
- Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–1162. - PubMed
-
- Pâques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. 2007;7:49–66. - PubMed
-
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotech. 2005;23:967–973. - PubMed
-
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. - PubMed
Publication types
MeSH terms
Grants and funding
- UL1 RR024921-01/RR/NCRR NIH HHS/United States
- RL1 CA133832-03/CA/NCI NIH HHS/United States
- R01 HL075453/HL/NHLBI NIH HHS/United States
- T32 GM07270/GM/NIGMS NIH HHS/United States
- UL1 DE019582-02/DE/NIDCR NIH HHS/United States
- RL1 CA133832-02/CA/NCI NIH HHS/United States
- T32 GM007270/GM/NIGMS NIH HHS/United States
- PL1-HL092557/HL/NHLBI NIH HHS/United States
- R21 AI064581-02/AI/NIAID NIH HHS/United States
- RL1CA133832/CA/NCI NIH HHS/United States
- R01-HL075453/HL/NHLBI NIH HHS/United States
- UL1 RR024921/RR/NCRR NIH HHS/United States
- RL1-HL092553/HL/NHLBI NIH HHS/United States
- UL1DE019582/DE/NIDCR NIH HHS/United States
- RL1 HL092553/HL/NHLBI NIH HHS/United States
- UL1 DE019582-03/DE/NIDCR NIH HHS/United States
- RL1 CA133832/CA/NCI NIH HHS/United States
- PL1 HL092557/HL/NHLBI NIH HHS/United States
- RL1 CA133832-01/CA/NCI NIH HHS/United States
- R21 AI064581/AI/NIAID NIH HHS/United States
- UL1 DE019582/DE/NIDCR NIH HHS/United States
- UL1 DE019582-05/DE/NIDCR NIH HHS/United States
- UL1 DE019582-04/DE/NIDCR NIH HHS/United States
- RL1 CA133832-04/CA/NCI NIH HHS/United States
- RL1 CA133832-05/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

