Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection
- PMID: 21742798
- PMCID: PMC3205871
- DOI: 10.1074/mcp.M111.009936
Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection
Abstract
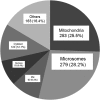
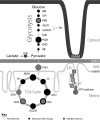
Endoplasmic reticulum-mitochondrial contacts, known as mitochondria-associated membranes, regulate important cellular functions including calcium signaling, bioenergetics, and apoptosis. Human cytomegalovirus is a medically important herpesvirus whose growth increases energy demand and depends upon continued cell survival. To gain insight into how human cytomegalovirus infection affects endoplasmic reticulum-mitochondrial contacts, we undertook quantitative proteomics of mitochondria-associated membranes using differential stable isotope labeling by amino acids in cell culture strategy and liquid chromatography-tandem MS analysis. This is the first reported quantitative proteomic analyses of a suborganelle during permissive human cytomegalovirus infection. Human fibroblasts were uninfected or human cytomegalovirus-infected for 72 h. Heavy mitochondria-associated membranes were isolated from paired unlabeled, uninfected cells and stable isotope labeling by amino acids in cell culture-labeled, infected cells and analyzed by liquid chromatography-tandem MS analysis. The results were verified by a reverse labeling experiment. Human cytomegalovirus infection dramatically altered endoplasmic reticulum-mitochondrial contacts by late times. Notable is the increased abundance of several fundamental networks in the mitochondria-associated membrane fraction of human cytomegalovirus-infected fibroblasts. Chaperones, including HSP60 and BiP, which is required for human cytomegalovirus assembly, were prominently increased at endoplasmic reticulum-mitochondrial contacts after infection. Minimal translational and translocation machineries were also associated with endoplasmic reticulum-mitochondrial contacts and increased after human cytomegalovirus infection as were glucose regulated protein 75 and the voltage dependent anion channel, which can form an endoplasmic reticulum-mitochondrial calcium signaling complex. Surprisingly, mitochondrial metabolic enzymes and cytosolic glycolytic enzymes were confidently detected in the mitochondria-associated membrane fraction and increased therein after infection. Finally, proapoptotic regulatory proteins, including Bax, cytochrome c, and Opa1, were augmented in endoplasmic reticulum-mitochondrial contacts after infection, suggesting attenuation of proapoptotic signaling by their increased presence therein. Together, these results suggest that human cytomegalovirus infection restructures the proteome of endoplasmic reticulum-mitochondrial contacts to bolster protein translation at these junctions, calcium signaling to mitochondria, cell survival, and bioenergetics and, thereby, allow for enhanced progeny production.
Figures
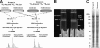

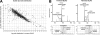
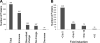


Similar articles
-
HCMV strain- and cell type-specific alterations in membrane contact sites point to the convergent regulation of organelle remodeling.J Virol. 2024 Nov 19;98(11):e0109924. doi: 10.1128/jvi.01099-24. Epub 2024 Oct 31. J Virol. 2024. PMID: 39480111
-
Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum-mitochondrial contacts.J Biol Chem. 2017 Sep 29;292(39):16382-16392. doi: 10.1074/jbc.M117.795286. Epub 2017 Jul 31. J Biol Chem. 2017. PMID: 28760823 Free PMC article.
-
Superresolution imaging of viral protein trafficking.Med Microbiol Immunol. 2015 Jun;204(3):449-60. doi: 10.1007/s00430-015-0395-0. Epub 2015 Feb 28. Med Microbiol Immunol. 2015. PMID: 25724304 Free PMC article. Review.
-
Plastic mitochondria-endoplasmic reticulum (ER) contacts use chaperones and tethers to mould their structure and signaling.Curr Opin Cell Biol. 2018 Aug;53:61-69. doi: 10.1016/j.ceb.2018.04.014. Epub 2018 Jun 2. Curr Opin Cell Biol. 2018. PMID: 29870872 Review.
-
Chemical Profiling of the Endoplasmic Reticulum Proteome Using Designer Labeling Reagents.J Am Chem Soc. 2018 Dec 12;140(49):17060-17070. doi: 10.1021/jacs.8b08606. Epub 2018 Nov 28. J Am Chem Soc. 2018. PMID: 30433779
Cited by
-
Mitochondria-endoplasmic reticulum contacts in sepsis-induced myocardial dysfunction.Front Cell Dev Biol. 2022 Nov 24;10:1036225. doi: 10.3389/fcell.2022.1036225. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506093 Free PMC article. Review.
-
Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation.Elife. 2017 Apr 25;6:e24463. doi: 10.7554/eLife.24463. Elife. 2017. PMID: 28441135 Free PMC article.
-
BCL2L13 at endoplasmic reticulum-mitochondria contact sites regulates calcium homeostasis to maintain skeletal muscle function.iScience. 2024 Jul 14;27(8):110510. doi: 10.1016/j.isci.2024.110510. eCollection 2024 Aug 16. iScience. 2024. PMID: 39175772 Free PMC article.
-
IRES-Dependent Translational Control during Virus-Induced Endoplasmic Reticulum Stress and Apoptosis.Front Microbiol. 2012 Mar 19;3:92. doi: 10.3389/fmicb.2012.00092. eCollection 2012. Front Microbiol. 2012. PMID: 22461781 Free PMC article.
-
The Role of Mitochondria in Inflammation: From Cancer to Neurodegenerative Disorders.J Clin Med. 2020 Mar 9;9(3):740. doi: 10.3390/jcm9030740. J Clin Med. 2020. PMID: 32182899 Free PMC article. Review.
References
-
- Mocarski E. S., Shenk T., Pass R. F. (2007) Cytomegaloviruses. In: Knipe D. M., Howley P. M. eds. Fields Virology, 5th Ed., pp. 2701–2772, Wolters Kluwer Health, Lippincott Williams & Wilkins, Philadelphia
-
- Gerna G., Baldanti F., Revello M. G. (2004) Pathogenesis of human cytomegalovirus infection and cellular targets. Hum. Immunol. 65, 381–386 - PubMed
-
- Boeckh M., Nichols W. G. (2004) The impact of cytomegalovirus serostatus of donor and recipient before hematopoietic stem cell transplantation in the era of antiviral prophylaxis and preemptive therapy. Blood. 103, 2003–2008 - PubMed
-
- Khanna R., Diamond D. J. (2006) Human cytomegalovirus vaccine: time to look for alternative options. Trends Mol. Med. 12, 26–33 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

