ClpXP, an ATP-powered unfolding and protein-degradation machine
- PMID: 21736903
- PMCID: PMC3209554
- DOI: 10.1016/j.bbamcr.2011.06.007
ClpXP, an ATP-powered unfolding and protein-degradation machine
Abstract
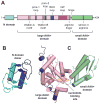
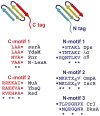
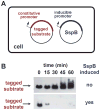
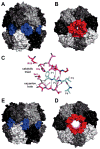
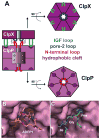
ClpXP is a AAA+ protease that uses the energy of ATP binding and hydrolysis to perform mechanical work during targeted protein degradation within cells. ClpXP consists of hexamers of a AAA+ ATPase (ClpX) and a tetradecameric peptidase (ClpP). Asymmetric ClpX hexamers bind unstructured peptide tags in protein substrates, unfold stable tertiary structure in the substrate, and then translocate the unfolded polypeptide chain into an internal proteolytic compartment in ClpP. Here, we review our present understanding of ClpXP structure and function, as revealed by two decades of biochemical and biophysical studies.
Copyright © 2011 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.Elife. 2020 Jan 9;9:e52158. doi: 10.7554/eLife.52158. Elife. 2020. PMID: 31916936 Free PMC article.
-
Hinge-Linker Elements in the AAA+ Protein Unfoldase ClpX Mediate Intersubunit Communication, Assembly, and Mechanical Activity.Biochemistry. 2018 Dec 11;57(49):6787-6796. doi: 10.1021/acs.biochem.8b00907. Epub 2018 Nov 20. Biochemistry. 2018. PMID: 30418765 Free PMC article.
-
Slippery substrates impair function of a bacterial protease ATPase by unbalancing translocation versus exit.J Biol Chem. 2013 May 10;288(19):13243-57. doi: 10.1074/jbc.M113.452524. Epub 2013 Mar 25. J Biol Chem. 2013. PMID: 23530043 Free PMC article.
-
Structure and function of ClpXP, a AAA+ proteolytic machine powered by probabilistic ATP hydrolysis.Crit Rev Biochem Mol Biol. 2022 Apr;57(2):188-204. doi: 10.1080/10409238.2021.1979461. Epub 2021 Dec 19. Crit Rev Biochem Mol Biol. 2022. PMID: 34923891 Free PMC article. Review.
-
Progress and prospect of single-molecular ClpX ATPase researching system-a mini-review.Gene. 2021 Mar 30;774:145420. doi: 10.1016/j.gene.2021.145420. Epub 2021 Jan 9. Gene. 2021. PMID: 33434627 Review.
Cited by
-
Structural insights into the inactive subunit of the apicoplast-localized caseinolytic protease complex of Plasmodium falciparum.J Biol Chem. 2013 Jan 11;288(2):1022-31. doi: 10.1074/jbc.M112.416560. Epub 2012 Nov 28. J Biol Chem. 2013. PMID: 23192353 Free PMC article.
-
Tag-Dependent Substrate Selection of ClpX Underlies Secondary Differentiation of Chlamydia trachomatis.mBio. 2022 Oct 26;13(5):e0185822. doi: 10.1128/mbio.01858-22. Epub 2022 Sep 26. mBio. 2022. PMID: 36154190 Free PMC article.
-
Sec18 side-loading is essential for universal SNARE recycling across cellular contexts.bioRxiv [Preprint]. 2024 Sep 1:2024.08.30.610324. doi: 10.1101/2024.08.30.610324. bioRxiv. 2024. PMID: 39257774 Free PMC article. Preprint.
-
Substrate-translocating loops regulate mechanochemical coupling and power production in AAA+ protease ClpXP.Nat Struct Mol Biol. 2016 Nov;23(11):974-981. doi: 10.1038/nsmb.3298. Epub 2016 Sep 26. Nat Struct Mol Biol. 2016. PMID: 27669037 Free PMC article.
-
CRISPR Interference To Inducibly Repress Gene Expression in Chlamydia trachomatis.Infect Immun. 2021 Jun 16;89(7):e0010821. doi: 10.1128/IAI.00108-21. Epub 2021 Jun 16. Infect Immun. 2021. PMID: 33875479 Free PMC article.
References
-
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. - PubMed
-
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011 Jun 23; [Epub ahead of print] - PubMed
-
- Katayama-Fujimura Y, Gottesman S, Maurizi MR. A multiple-component, ATP-dependent protease from Escherichia coli. J Biol Chem. 1987;262:4477–4485. - PubMed
-
- Katayama Y, Gottesman S, Pumphrey J, Rudikoff S, Clark WP, Maurizi MR. The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning, and mutational analysis of the ATP-binding component. J Biol Chem. 1988;263:15226–15236. - PubMed
-
- Hwang BJ, Woo KM, Goldberg AL, Chung CH. Protease Ti, a new ATP-dependent protease in Escherichia coli,contains protein-activated ATPase and proteolytic functions in distinct subunits. J Biol Chem. 1988;263:8727–8734. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AI015706-33/AI/NIAID NIH HHS/United States
- R37 AI015706-32/AI/NIAID NIH HHS/United States
- R01 AI016892/AI/NIAID NIH HHS/United States
- R01 AI016892-33/AI/NIAID NIH HHS/United States
- R01 AI016892-29/AI/NIAID NIH HHS/United States
- R01 AI016892-32/AI/NIAID NIH HHS/United States
- R01 AI016892-30/AI/NIAID NIH HHS/United States
- R01 AI016892-31/AI/NIAID NIH HHS/United States
- R01 AI015706/AI/NIAID NIH HHS/United States
- R37 AI015706/AI/NIAID NIH HHS/United States
- R01 GM101988/GM/NIGMS NIH HHS/United States
- AI-15706/AI/NIAID NIH HHS/United States
- GM-499224/GM/NIGMS NIH HHS/United States
- R37 AI015706-30/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R37 AI015706-29/AI/NIAID NIH HHS/United States
- AI-16892/AI/NIAID NIH HHS/United States
- R37 AI015706-31/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

