Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain
- PMID: 21731699
- PMCID: PMC3120858
- DOI: 10.1371/journal.pone.0021306
Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain
Abstract
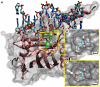
Recent discovery of 5-hydroxymethylcytosine (5hmC) in genomic DNA raises the question how this sixth base is recognized by cellular proteins. In contrast to the methyl-CpG binding domain (MBD) of MeCP2, we found that the SRA domain of Uhrf1, an essential factor in DNA maintenance methylation, binds 5hmC and 5-methylcytosine containing substrates with similar affinity. Based on the co-crystal structure, we performed molecular dynamics simulations of the SRA:DNA complex with the flipped cytosine base carrying either of these epigenetic modifications. Our data indicate that the SRA binding pocket can accommodate 5hmC and stabilizes the flipped base by hydrogen bond formation with the hydroxyl group.
Conflict of interest statement
Figures
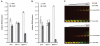

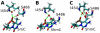
Similar articles
-
Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.Nature. 2008 Oct 9;455(7214):822-5. doi: 10.1038/nature07273. Epub 2008 Sep 3. Nature. 2008. PMID: 18772889
-
Systematic analysis of the binding behaviour of UHRF1 towards different methyl- and carboxylcytosine modification patterns at CpG dyads.PLoS One. 2020 Feb 21;15(2):e0229144. doi: 10.1371/journal.pone.0229144. eCollection 2020. PLoS One. 2020. PMID: 32084194 Free PMC article.
-
UHRF1 discriminates against binding to fully-methylated CpG-Sites by steric repulsion.Biophys Chem. 2013 Jan;171:38-45. doi: 10.1016/j.bpc.2012.10.002. Epub 2012 Oct 22. Biophys Chem. 2013. PMID: 23245651
-
Novel DNA binding domain-based assays for detection of methylated and nonmethylated DNA.Epigenomics. 2011 Feb;3(1):93-101. doi: 10.2217/epi.10.69. Epigenomics. 2011. PMID: 22126156 Review.
-
Genomic distribution and possible functions of DNA hydroxymethylation in the brain.Genomics. 2014 Nov;104(5):341-6. doi: 10.1016/j.ygeno.2014.08.020. Epub 2014 Sep 7. Genomics. 2014. PMID: 25205307 Review.
Cited by
-
TET proteins: on the frenetic hunt for new cytosine modifications.Brief Funct Genomics. 2013 May;12(3):191-204. doi: 10.1093/bfgp/elt010. Epub 2013 Apr 26. Brief Funct Genomics. 2013. PMID: 23625996 Free PMC article. Review.
-
Role of TET enzymes in DNA methylation, development, and cancer.Genes Dev. 2016 Apr 1;30(7):733-50. doi: 10.1101/gad.276568.115. Genes Dev. 2016. PMID: 27036965 Free PMC article. Review.
-
Mapping recently identified nucleotide variants in the genome and transcriptome.Nat Biotechnol. 2012 Nov;30(11):1107-16. doi: 10.1038/nbt.2398. Nat Biotechnol. 2012. PMID: 23138310 Free PMC article. Review.
-
DNA recognition of 5-carboxylcytosine by a Zfp57 mutant at an atomic resolution of 0.97 Å.Biochemistry. 2013 Dec 23;52(51):9310-7. doi: 10.1021/bi401360n. Epub 2013 Nov 20. Biochemistry. 2013. PMID: 24236546 Free PMC article.
-
The curious chemical biology of cytosine: deamination, methylation, and oxidation as modulators of genomic potential.ACS Chem Biol. 2012 Jan 20;7(1):20-30. doi: 10.1021/cb2002895. Epub 2011 Oct 31. ACS Chem Biol. 2012. PMID: 22004246 Free PMC article. Review.
References
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Rottach A, Leonhardt H, Spada F. DNA methylation-mediated epigenetic control. J Cell Biochem. 2009;108:43–51. - PubMed
-
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
-
- Lei H, Oh S, Okano M, Juttermann R, Goss K, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. - PubMed
-
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

