Single-molecule investigations of the stringent response machinery in living bacterial cells
- PMID: 21730169
- PMCID: PMC3150888
- DOI: 10.1073/pnas.1102255108
Single-molecule investigations of the stringent response machinery in living bacterial cells
Abstract
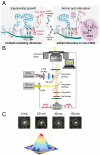
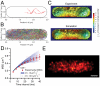
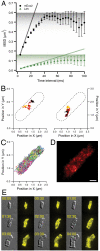
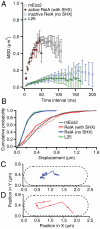
The RelA-mediated stringent response is at the heart of bacterial adaptation to starvation and stress, playing a major role in the bacterial cell cycle and virulence. RelA integrates several environmental cues and synthesizes the alarmone ppGpp, which globally reprograms transcription, translation, and replication. We have developed and implemented novel single-molecule tracking methodology to characterize the intracellular catalytic cycle of RelA. Our single-molecule experiments show that RelA is on the ribosome under nonstarved conditions and that the individual enzyme molecule stays off the ribosome for an extended period of time after activation. This suggests that the catalytically active part of the RelA cycle is performed off, rather than on, the ribosome, and that rebinding to the ribosome is not necessary to trigger each ppGpp synthesis event. Furthermore, we find fast activation of RelA in response to heat stress followed by RelA rapidly being reset to its inactive state, which makes the system sensitive to new environmental cues and hints at an underlying excitable response mechanism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Genetic analysis of the invariant residue G791 in Escherichia coli 16S rRNA implicates RelA in ribosome function.J Bacteriol. 2009 Apr;191(7):2042-50. doi: 10.1128/JB.00904-08. Epub 2009 Jan 23. J Bacteriol. 2009. PMID: 19168615 Free PMC article.
-
Positive allosteric feedback regulation of the stringent response enzyme RelA by its product.EMBO Rep. 2012 Sep;13(9):835-9. doi: 10.1038/embor.2012.106. Epub 2012 Jul 20. EMBO Rep. 2012. PMID: 22814757 Free PMC article.
-
Ribosome-dependent activation of stringent control.Nature. 2016 Jun 9;534(7606):277-280. doi: 10.1038/nature17675. Epub 2016 May 9. Nature. 2016. PMID: 27279228 Free PMC article.
-
How to save a bacterial ribosome in times of stress.Semin Cell Dev Biol. 2023 Feb 28;136:3-12. doi: 10.1016/j.semcdb.2022.03.015. Epub 2022 Mar 21. Semin Cell Dev Biol. 2023. PMID: 35331628 Review.
-
Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants.Plant Biol (Stuttg). 2011 Sep;13(5):699-709. doi: 10.1111/j.1438-8677.2011.00484.x. Epub 2011 May 31. Plant Biol (Stuttg). 2011. PMID: 21815973 Review.
Cited by
-
Real-time single-molecule imaging of transcriptional regulatory networks in living cells.Nat Rev Genet. 2024 Apr;25(4):272-285. doi: 10.1038/s41576-023-00684-9. Epub 2024 Jan 9. Nat Rev Genet. 2024. PMID: 38195868 Review.
-
In vivo architecture and action of bacterial structural maintenance of chromosome proteins.Science. 2012 Oct 26;338(6106):528-31. doi: 10.1126/science.1227126. Science. 2012. PMID: 23112333 Free PMC article.
-
Single-Molecule Tracking and Its Application in Biomolecular Binding Detection.IEEE J Sel Top Quantum Electron. 2016 Jul-Aug;22(4):6804013. doi: 10.1109/JSTQE.2016.2568160. Epub 2016 May 17. IEEE J Sel Top Quantum Electron. 2016. PMID: 27660404 Free PMC article.
-
RsfA (YbeB) proteins are conserved ribosomal silencing factors.PLoS Genet. 2012;8(7):e1002815. doi: 10.1371/journal.pgen.1002815. Epub 2012 Jul 19. PLoS Genet. 2012. PMID: 22829778 Free PMC article.
-
Simulation-based Reconstructed Diffusion unveils the effect of aging on protein diffusion in Escherichia coli.PLoS Comput Biol. 2023 Sep 11;19(9):e1011093. doi: 10.1371/journal.pcbi.1011093. eCollection 2023 Sep. PLoS Comput Biol. 2023. PMID: 37695774 Free PMC article.
References
-
- Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

