Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects
- PMID: 21730157
- PMCID: PMC3141927
- DOI: 10.1073/pnas.1100923108
Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects
Abstract
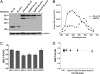
Identifying protein-protein interactions (PPIs) is essential for understanding various disease mechanisms and developing new therapeutic approaches. Current methods for assaying cellular intermolecular interactions are mainly used for cells in culture and have limited use for the noninvasive assessment of small animal disease models. Here, we describe red light-emitting reporter systems based on bioluminescence resonance energy transfer (BRET) that allow for assaying PPIs both in cell culture and deep tissues of small animals. These BRET systems consist of the recently developed Renilla reniformis luciferase (RLuc) variants RLuc8 and RLuc8.6, used as BRET donors, combined with two red fluorescent proteins, TagRFP and TurboFP635, as BRET acceptors. In addition to the native coelenterazine luciferase substrate, we used the synthetic derivative coelenterazine-v, which further red-shifts the emission maxima of Renilla luciferases by 35 nm. We show the use of these BRET systems for ratiometric imaging of both cells in culture and deep-tissue small animal tumor models and validate their applicability for studying PPIs in mice in the context of rapamycin-induced FK506 binding protein 12 (FKBP12)-FKBP12 rapamycin binding domain (FRB) association. These red light-emitting BRET systems have great potential for investigating PPIs in the context of drug screening and target validation applications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
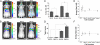
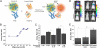
Similar articles
-
BRET3: a red-shifted bioluminescence resonance energy transfer (BRET)-based integrated platform for imaging protein-protein interactions from single live cells and living animals.FASEB J. 2009 Aug;23(8):2702-9. doi: 10.1096/fj.08-118919. Epub 2009 Apr 7. FASEB J. 2009. PMID: 19351700 Free PMC article.
-
An improved bioluminescence resonance energy transfer strategy for imaging intracellular events in single cells and living subjects.Cancer Res. 2007 Aug 1;67(15):7175-83. doi: 10.1158/0008-5472.CAN-06-4623. Cancer Res. 2007. PMID: 17671185 Free PMC article.
-
Ligand-activated BRET9 imaging for measuring protein-protein interactions in living mice.Chem Commun (Camb). 2019 Dec 19;56(2):281-284. doi: 10.1039/c9cc07634d. Chem Commun (Camb). 2019. PMID: 31807738
-
Biosensing and imaging based on bioluminescence resonance energy transfer.Curr Opin Biotechnol. 2009 Feb;20(1):37-44. doi: 10.1016/j.copbio.2009.01.001. Epub 2009 Feb 11. Curr Opin Biotechnol. 2009. PMID: 19216068 Free PMC article. Review.
-
The BRET technology and its application to screening assays.Biotechnol J. 2008 Mar;3(3):311-24. doi: 10.1002/biot.200700222. Biotechnol J. 2008. PMID: 18228541 Review.
Cited by
-
Advanced Bioluminescence System for In Vivo Imaging with Brighter and Red-Shifted Light Emission.Int J Mol Sci. 2020 Sep 7;21(18):6538. doi: 10.3390/ijms21186538. Int J Mol Sci. 2020. PMID: 32906768 Free PMC article. Review.
-
Bioluminescence: a versatile technique for imaging cellular and molecular features.Medchemcomm. 2014 Mar 1;5(3):255-267. doi: 10.1039/C3MD00288H. Epub 2013 Dec 13. Medchemcomm. 2014. PMID: 27594981 Free PMC article.
-
A novel injectable BRET-based in vivo imaging probe for detecting the activity of hypoxia-inducible factor regulated by the ubiquitin-proteasome system.Sci Rep. 2016 Oct 4;6:34311. doi: 10.1038/srep34311. Sci Rep. 2016. PMID: 27698477 Free PMC article.
-
Sensitive and high resolution localization and tracking of membrane proteins in live cells with BRET.Traffic. 2012 Nov;13(11):1450-6. doi: 10.1111/j.1600-0854.2012.01401.x. Epub 2012 Aug 5. Traffic. 2012. PMID: 22816793 Free PMC article.
-
Mathematical models for quantitative assessment of bioluminescence resonance energy transfer: application to seven transmembrane receptors oligomerization.Front Endocrinol (Lausanne). 2012 Aug 28;3:104. doi: 10.3389/fendo.2012.00104. eCollection 2012. Front Endocrinol (Lausanne). 2012. PMID: 22973259 Free PMC article.
References
-
- Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. - PubMed
-
- Guan H, Kiss-Toth E. Advanced technologies for studies on protein interactomes. Adv Biochem Eng Biotechnol. 2008;110:1–24. - PubMed
-
- Luker GD, Sharma V, Piwnica-Worms D. Visualizing protein-protein interactions in living animals. Methods. 2003;29:110–122. - PubMed
-
- Truong K, Ikura M. The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo. Curr Opin Struct Biol. 2001;11:573–578. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

