Reductive evolution of proteomes and protein structures
- PMID: 21730144
- PMCID: PMC3141956
- DOI: 10.1073/pnas.1017361108
Reductive evolution of proteomes and protein structures
Abstract
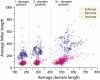
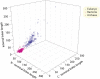
The lengths of orthologous protein families in Eukarya are almost double the lengths found in Bacteria and Archaea. Here we examine protein structures in 745 genomes and show that protein length differences between superkingdoms arise as much shorter prokaryotic nondomain linker sequences. Eukaryotic, bacterial, and archaeal linkers are 250, 86, and 73 aa residues in length, respectively, whereas folded domain sequences are 281, 280, and 256 residues, respectively. Cryptic domains match linkers (P < 0.0001) with probabilities ranging between 0.022 and 0.042; accordingly, they do not affect length estimates significantly. Linker sequences support intermolecular binding within proteomes and they are probably enriched in intrinsically disordered regions as well. Reductively evolved linker sequence lengths in growth rate maximized cells should be proportional to proteome diversity. By using total in-frame coding capacity of a genome [i.e., coding sequence (CDS)] as a reliable measure of proteome diversity, we find linker lengths of prokaryotes clearly evolve in proportion to CDS values, whereas those of eukaryotes are more randomly larger than expected. Domain lengths scarcely change over the entire range of CDS values. Thus, the protein linkers of prokaryotes evolve reductively whereas those of eukaryotes do not.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The origins of modern proteomes.Biochimie. 2007 Dec;89(12):1454-63. doi: 10.1016/j.biochi.2007.09.004. Epub 2007 Sep 15. Biochimie. 2007. PMID: 17949885
-
The evolutionary history of protein fold families and proteomes confirms that the archaeal ancestor is more ancient than the ancestors of other superkingdoms.BMC Evol Biol. 2012 Jan 27;12:13. doi: 10.1186/1471-2148-12-13. BMC Evol Biol. 2012. PMID: 22284070 Free PMC article.
-
Global patterns of protein domain gain and loss in superkingdoms.PLoS Comput Biol. 2014 Jan 30;10(1):e1003452. doi: 10.1371/journal.pcbi.1003452. eCollection 2014 Jan. PLoS Comput Biol. 2014. PMID: 24499935 Free PMC article.
-
Comparative Genomics for Prokaryotes.Methods Mol Biol. 2018;1704:55-78. doi: 10.1007/978-1-4939-7463-4_3. Methods Mol Biol. 2018. PMID: 29277863 Review.
-
[Homologous protein domains in superkingdoms Archaea, Bacteria, and Eukaryota and the problem of the origin of eukaryotes].Izv Akad Nauk Ser Biol. 2005 Jul-Aug;(4):389-400. Izv Akad Nauk Ser Biol. 2005. PMID: 16212260 Review. Russian.
Cited by
-
The organization of domains in proteins obeys Menzerath-Altmann's law of language.BMC Syst Biol. 2015 Aug 11;9:44. doi: 10.1186/s12918-015-0192-9. BMC Syst Biol. 2015. PMID: 26260760 Free PMC article.
-
Annotation of Protein Domains Reveals Remarkable Conservation in the Functional Make up of Proteomes Across Superkingdoms.Genes (Basel). 2011 Nov 8;2(4):869-911. doi: 10.3390/genes2040869. Genes (Basel). 2011. PMID: 24710297 Free PMC article.
-
The Compressed Vocabulary of Microbial Life.Front Microbiol. 2021 Jul 7;12:655990. doi: 10.3389/fmicb.2021.655990. eCollection 2021. Front Microbiol. 2021. PMID: 34305827 Free PMC article.
-
Characterization of Three L-Asparaginases from Maritime Pine (Pinus pinaster Ait.).Front Plant Sci. 2017 Jun 23;8:1075. doi: 10.3389/fpls.2017.01075. eCollection 2017. Front Plant Sci. 2017. PMID: 28690619 Free PMC article.
-
The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis.Cell. 2013 Jan 17;152(1-2):196-209. doi: 10.1016/j.cell.2012.12.001. Cell. 2013. PMID: 23332755 Free PMC article.
References
-
- Zhang J. Protein-length distributions for the three domains of life. Trends Genet. 2000;16:107–109. - PubMed
-
- Liang P, Riley M. A comparative genomics approach for studying ancestral proteins and evolution. Adv Appl Microbiol. 2001;50:39–72. - PubMed
-
- Kurland CG, Canbäck B, Berg OG. The origins of modern proteomes. Biochimie. 2007;89:1454–1463. - PubMed
-
- Ehrenberg M, Kurland CG. Costs of accuracy determined by a maximal growth rate constraint. Q Rev Biophys. 1984;17:45–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

