The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes
- PMID: 21725321
- PMCID: PMC3324395
- DOI: 10.1038/ni.2063
The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes
Abstract
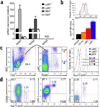
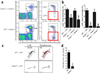
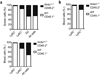
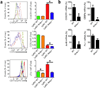
The transcription factors that regulate differentiation into the monocyte subset in bone marrow have not yet been identified. Here we found that the orphan nuclear receptor NR4A1 controlled the differentiation of Ly6C- monocytes. Ly6C- monocytes, which function in a surveillance role in circulation, were absent from Nr4a1-/- mice. Normal numbers of myeloid progenitor cells were present in Nr4a1-/- mice, which indicated that the defect occurred during later stages of monocyte development. The defect was cell intrinsic, as wild-type mice that received bone marrow from Nr4a1-/- mice developed fewer patrolling monocytes than did recipients of wild-type bone marrow. The Ly6C- monocytes remaining in the bone marrow of Nr4a1-/- mice were arrested in S phase of the cell cycle and underwent apoptosis. Thus, NR4A1 functions as a master regulator of the differentiation and survival of 'patrolling' Ly6C- monocytes.
Figures




Comment in
-
NUR who? An orphan transcription factor holds promise for monomaniacs.Nat Immunol. 2011 Jul 19;12(8):727-9. doi: 10.1038/ni.2074. Nat Immunol. 2011. PMID: 21772283 No abstract available.
Similar articles
-
Deleting an Nr4a1 Super-Enhancer Subdomain Ablates Ly6Clow Monocytes while Preserving Macrophage Gene Function.Immunity. 2016 Nov 15;45(5):975-987. doi: 10.1016/j.immuni.2016.10.011. Epub 2016 Nov 1. Immunity. 2016. PMID: 27814941 Free PMC article.
-
Ly6C- Monocytes Regulate Parasite-Induced Liver Inflammation by Inducing the Differentiation of Pathogenic Ly6C+ Monocytes into Macrophages.PLoS Pathog. 2015 May 28;11(5):e1004873. doi: 10.1371/journal.ppat.1004873. eCollection 2015 May. PLoS Pathog. 2015. PMID: 26020782 Free PMC article.
-
NR4A1-dependent Ly6Clow monocytes contribute to reducing joint inflammation in arthritic mice through Treg cells.Eur J Immunol. 2016 Dec;46(12):2789-2800. doi: 10.1002/eji.201646406. Epub 2016 Oct 18. Eur J Immunol. 2016. PMID: 27600773
-
The nuclear orphan receptor NR4A1 and NR4A3 as tumor suppressors in hematologic neoplasms.Curr Drug Targets. 2015;16(1):38-46. doi: 10.2174/1389450115666141120112818. Curr Drug Targets. 2015. PMID: 25410408 Review.
-
Monocyte-mediated immune defense against murine Listeria monocytogenes infection.Adv Immunol. 2012;113:119-34. doi: 10.1016/B978-0-12-394590-7.00003-8. Adv Immunol. 2012. PMID: 22244581 Free PMC article. Review.
Cited by
-
Unravelling monocyte functions: from the guardians of health to the regulators of disease.Discov Immunol. 2024 Aug 30;3(1):kyae014. doi: 10.1093/discim/kyae014. eCollection 2024. Discov Immunol. 2024. PMID: 39430099 Free PMC article. Review.
-
Immunological Feature and Transcriptional Signaling of Ly6C Monocyte Subsets From Transcriptome Analysis in Control and Hyperhomocysteinemic Mice.Front Immunol. 2021 Feb 25;12:632333. doi: 10.3389/fimmu.2021.632333. eCollection 2021. Front Immunol. 2021. PMID: 33717169 Free PMC article.
-
Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation.Blood. 2013 Mar 7;121(10):1839-49. doi: 10.1182/blood-2012-06-437863. Epub 2013 Jan 14. Blood. 2013. PMID: 23319570 Free PMC article.
-
The dynamics of monocytes and microglia in Alzheimer's disease.Alzheimers Res Ther. 2015 Apr 15;7(1):41. doi: 10.1186/s13195-015-0125-2. eCollection 2015. Alzheimers Res Ther. 2015. PMID: 25878730 Free PMC article.
-
Elucidation of monocyte/macrophage dynamics and function by intravital imaging.J Leukoc Biol. 2015 Sep;98(3):319-32. doi: 10.1189/jlb.4RI0115-006RR. Epub 2015 Jul 10. J Leukoc Biol. 2015. PMID: 26162402 Free PMC article. Review.
References
-
- Martínez-González J, Badimon L. The NR4A subfamily of nuclear receptors: new early genes regulated by growth factors in vascular cells. Cardiovasc Res. 2005;65:609–618. - PubMed
-
- Lim RW, Varnum BC, Herschman HR. Cloning of tetradecanoyl phorbol ester-induced 'primary response' sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene. 1987;1:263–270. - PubMed
-
- Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3 - transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–4743. - PubMed
-
- Li QX, Ke N, Sundaram R, Wong-Staal F. NR4A1, 2, 3--an orphan nuclear hormone receptor family involved in cell apoptosis and carcinogenesis. Histol Histopathol. 2006;21:533–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

