Allosteric interactions across native adenosine-A3 receptor homodimers: quantification using single-cell ligand-binding kinetics
- PMID: 21715680
- PMCID: PMC3177574
- DOI: 10.1096/fj.11-186296
Allosteric interactions across native adenosine-A3 receptor homodimers: quantification using single-cell ligand-binding kinetics
Abstract
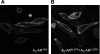
A growing awareness indicates that many G-protein-coupled receptors (GPCRs) exist as homodimers, but the extent of the cooperativity across the dimer interface has been largely unexplored. Here, measurement of the dissociation kinetics of a fluorescent agonist (ABA-X-BY630) from the human A(1) or A(3) adenosine receptors expressed in CHO-K1 cells has provided evidence for highly cooperative interactions between protomers of the A(3)-receptor dimer in single living cells. In the absence of competitive ligands, the dissociation rate constants of ABA-X-BY630 from A(1) and A(3) receptors were 1.45 ± 0.05 and 0.57 ± 0.07 min(-1), respectively. At the A(3) receptor, this could be markedly increased by both orthosteric agonists and antagonists [15-, 9-, and 19-fold for xanthine amine congener (XAC), 5'-(N-ethyl carboxamido)adenosine (NECA), and adenosine, respectively] and reduced by coexpression of a nonbinding (N250A) A(3)-receptor mutant. The changes in ABA-X-BY630 dissociation were much lower at the A(1) receptor (1.5-, 1.4-, and 1.5-fold). Analysis of the pEC(50) values of XAC, NECA, and adenosine for the ABA-X-BY630-occupied A(3)-receptor dimer yielded values of 6.0 ± 0.1, 5.9 ± 0.1, and 5.2 ± 0.1, respectively. This study provides new insight into the spatial and temporal specificity of drug action that can be provided by allosteric modulation across a GPCR homodimeric interface.
Figures
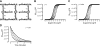


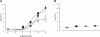
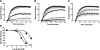

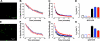
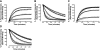
Similar articles
-
Antagonist selective modulation of adenosine A1 and A3 receptor pharmacology by the food dye Brilliant Black BN: evidence for allosteric interactions.Mol Pharmacol. 2010 Apr;77(4):678-86. doi: 10.1124/mol.109.063065. Epub 2010 Jan 19. Mol Pharmacol. 2010. PMID: 20086038 Free PMC article.
-
Direct visualisation of internalization of the adenosine A3 receptor and localization with arrestin3 using a fluorescent agonist.Neuropharmacology. 2015 Nov;98:68-77. doi: 10.1016/j.neuropharm.2015.04.013. Epub 2015 Apr 30. Neuropharmacology. 2015. PMID: 25937210
-
Different efficacy of adenosine and NECA derivatives at the human A3 adenosine receptor: insight into the receptor activation switch.Biochem Pharmacol. 2014 Jan 15;87(2):321-31. doi: 10.1016/j.bcp.2013.10.011. Epub 2013 Oct 23. Biochem Pharmacol. 2014. PMID: 24161786
-
Kinetic analysis of antagonist-occupied adenosine-A3 receptors within membrane microdomains of individual cells provides evidence of receptor dimerization and allosterism.FASEB J. 2014 Oct;28(10):4211-22. doi: 10.1096/fj.13-247270. Epub 2014 Jun 26. FASEB J. 2014. PMID: 24970394 Free PMC article.
-
Allosteric interactions at adenosine A(1) and A(3) receptors: new insights into the role of small molecules and receptor dimerization.Br J Pharmacol. 2014 Mar;171(5):1102-13. doi: 10.1111/bph.12345. Br J Pharmacol. 2014. PMID: 24024783 Free PMC article. Review.
Cited by
-
Migraine signaling pathways: purine metabolites that regulate migraine and predispose migraineurs to headache.Mol Cell Biochem. 2023 Dec;478(12):2813-2848. doi: 10.1007/s11010-023-04701-7. Epub 2023 Mar 22. Mol Cell Biochem. 2023. PMID: 36947357 Review.
-
Structural Probing and Molecular Modeling of the A₃ Adenosine Receptor: A Focus on Agonist Binding.Molecules. 2017 Mar 11;22(3):449. doi: 10.3390/molecules22030449. Molecules. 2017. PMID: 28287473 Free PMC article. Review.
-
Chemical Probes for the Adenosine Receptors.Pharmaceuticals (Basel). 2019 Nov 12;12(4):168. doi: 10.3390/ph12040168. Pharmaceuticals (Basel). 2019. PMID: 31726680 Free PMC article. Review.
-
Spatial Intensity Distribution Analysis: Studies of G Protein-Coupled Receptor Oligomerisation.Trends Pharmacol Sci. 2018 Feb;39(2):175-186. doi: 10.1016/j.tips.2017.09.001. Epub 2017 Oct 9. Trends Pharmacol Sci. 2018. PMID: 29032835 Free PMC article. Review.
-
Click modification in the N6 region of A3 adenosine receptor-selective carbocyclic nucleosides for dendrimeric tethering that preserves pharmacophore recognition.Bioconjug Chem. 2012 Feb 15;23(2):232-47. doi: 10.1021/bc200526c. Epub 2012 Jan 11. Bioconjug Chem. 2012. PMID: 22175234 Free PMC article.
References
-
- Overington J. P., Al-Lazikani B., Hopkins A. L. (2006) How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 - PubMed
-
- Kenakin T., Onaran O. (2002) The ligand paradox between affinity and efficacy: can you be there and not make a difference? Trends Pharmacol. Sci. 23, 275–280 - PubMed
-
- Birdsall N. J., Lazareno S. (2005) Allosterism at muscarinic receptors: ligands and mechanisms. Mini Rev. Med. Chem. 5, 523–543 - PubMed
-
- Christopoulos A. (2002) Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nat. Rev. Drug Discov. 1, 198–210 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

