The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection
- PMID: 21680511
- PMCID: PMC3165823
- DOI: 10.1128/JVI.02559-10
The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection
Abstract
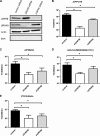
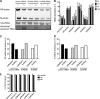
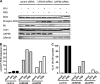
The cellular DEAD box RNA helicase UAP56 plays a pivotal role in the efficient transcription/replication of influenza A virus. UAP56 is recruited by the nucleoprotein (NP) of influenza A viruses, and recent data revealed that the RNA helicase is required for the nuclear export of a subset of spliced and unspliced viral mRNAs. The fact that influenza viruses do not produce detectable amounts of double-stranded RNA (dsRNA) intermediates during transcription/replication suggests the involvement of cellular RNA helicases. Hence, we examined whether the RNA-unwinding activity of UAP56 or its paralog URH49 plays a role in preventing the accumulation of dsRNA during infection. First, our data showed that not only UAP56 but also its paralog URH49 can interact with NPs of avian and human influenza A viruses. The small interfering RNA (siRNA)-mediated depletion of either RNA helicase reduced the transport of M1 and hemagglutinin (HA) mRNAs and, to a lesser extent, NP and NS1 mRNAs into the cytoplasm. Moreover, we found that virus infection of UAP56-depleted cells leads to the rapid accumulation of dsRNA in the perinuclear region. In parallel, we observed a robust virus-mediated activation of dsRNA-dependent protein kinase R (PKR), indicating that the cellular RNA helicase UAP56 may be recruited by influenza virus to prevent dsRNA formation. The accumulation of dsRNA was blocked when actinomycin D or cycloheximide was used to inhibit viral transcription/replication or translation, respectively. In summary, we demonstrate that UAP56 is utilized by influenza A viruses to prevent the formation of dsRNA and, hence, the activation of the innate immune response.
Figures
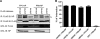

Similar articles
-
Interferon-induced antiviral protein MxA interacts with the cellular RNA helicases UAP56 and URH49.J Biol Chem. 2011 Oct 7;286(40):34743-51. doi: 10.1074/jbc.M111.251843. Epub 2011 Aug 22. J Biol Chem. 2011. PMID: 21859714 Free PMC article.
-
Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export.J Gen Virol. 2010 May;91(Pt 5):1290-301. doi: 10.1099/vir.0.018564-0. Epub 2010 Jan 13. J Gen Virol. 2010. PMID: 20071484 Free PMC article.
-
Cellular mRNA export factor UAP56 recognizes nucleic acid binding site of influenza virus NP protein.Biochem Biophys Res Commun. 2020 Apr 30;525(2):259-264. doi: 10.1016/j.bbrc.2020.02.059. Epub 2020 Feb 19. Biochem Biophys Res Commun. 2020. PMID: 32085897 Free PMC article.
-
Critical Cellular Functions and Mechanisms of Action of the RNA Helicase UAP56.J Mol Biol. 2024 Jun 15;436(12):168604. doi: 10.1016/j.jmb.2024.168604. Epub 2024 May 8. J Mol Biol. 2024. PMID: 38729260 Review.
-
Insight into Influenza: A Virus Cap-Snatching.Viruses. 2018 Nov 16;10(11):641. doi: 10.3390/v10110641. Viruses. 2018. PMID: 30453478 Free PMC article. Review.
Cited by
-
A new molecular classification to drive precision treatment strategies in primary Sjögren's syndrome.Nat Commun. 2021 Jun 10;12(1):3523. doi: 10.1038/s41467-021-23472-7. Nat Commun. 2021. PMID: 34112769 Free PMC article.
-
Quantitative proteomic analysis of the influenza A virus nonstructural proteins NS1 and NS2 during natural cell infection identifies PACT as an NS1 target protein and antiviral host factor.J Virol. 2014 Aug;88(16):9038-48. doi: 10.1128/JVI.00830-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899174 Free PMC article.
-
Protection from Severe Influenza Virus Infections in Mice Carrying the Mx1 Influenza Virus Resistance Gene Strongly Depends on Genetic Background.J Virol. 2015 Oct;89(19):9998-10009. doi: 10.1128/JVI.01305-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202236 Free PMC article.
-
Innate Immune Sensing of Influenza A Virus.Viruses. 2020 Jul 14;12(7):755. doi: 10.3390/v12070755. Viruses. 2020. PMID: 32674269 Free PMC article. Review.
-
Influenza A virus NS1 protein hijacks YAP/TAZ to suppress TLR3-mediated innate immune response.PLoS Pathog. 2022 May 3;18(5):e1010505. doi: 10.1371/journal.ppat.1010505. eCollection 2022 May. PLoS Pathog. 2022. PMID: 35503798 Free PMC article.
References
-
- Amorim M. J., Read E. K., Dalton R. M., Medcalf L., Digard P. 2007. Nuclear export of influenza A virus mRNAs requires ongoing RNA polymerase II activity. Traffic 8:1–11 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

