Four amino acids guide the assembly or disassembly of Arabidopsis histone H3.3-containing nucleosomes
- PMID: 21670303
- PMCID: PMC3127877
- DOI: 10.1073/pnas.1017882108
Four amino acids guide the assembly or disassembly of Arabidopsis histone H3.3-containing nucleosomes
Abstract
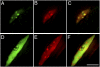
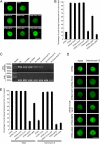
The histone variant H3.3 and the canonical histone H3.1, which differ in only 4- to 5-aa positions, are coexpressed in complex multicellular eukaryotes from fly to human and plant. H3.3 is mainly associated with active chromatin by replacing H3.1 through chaperones such as histone regulator A, death domain associated protein DAXX, thalassemia/mental retardation syndrome X-linked homolog ATRX, or proto-oncogene protein DEK and plays important roles in the germline, epigenetic memory, and reprogramming. However, the signals within H3.3 that serve as a guide for its dynamic deposition or depletion in plant chromatin are not clear. Here, we show that Arabidopsis histone H3.3 differs from H3.1 by 4-aa sites: amino acids 31, 41, 87, and 90. Although histone H3.1 is highly enriched in chromocenters, H3.3 is present in nucleolar foci in addition to being diffusely distributed in the nucleoplasm. We have evaluated the function of the 4 aa that differ between H3.1 and H3.3. We show that amino acid residue 87, and to some extent residue 90, of Arabidopsis histone H3.3 are critical for its deposition into rDNA arrays. When RNA polymerase I-directed nucleolar transcription is inhibited, wild type H3.3, but not H3.3 containing mutations at residues 31 and 41, is depleted from the rDNA arrays. Together, our results are consistent with a model in which amino acids 87 and 90 in the core domain of H3.3 guide nucleosome assembly, whereas amino acids 31 and 41 in the N-terminal tail of Arabidopsis H3.3 guide nucleosome disassembly in nucleolar rDNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements.Nucleic Acids Res. 2017 Jun 2;45(10):5691-5706. doi: 10.1093/nar/gkx131. Nucleic Acids Res. 2017. PMID: 28334823 Free PMC article.
-
Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres.Proc Natl Acad Sci U S A. 2010 Aug 10;107(32):14075-80. doi: 10.1073/pnas.1008850107. Epub 2010 Jul 22. Proc Natl Acad Sci U S A. 2010. PMID: 20651253 Free PMC article.
-
Arabidopsis ATRX Modulates H3.3 Occupancy and Fine-Tunes Gene Expression.Plant Cell. 2017 Jul;29(7):1773-1793. doi: 10.1105/tpc.16.00877. Epub 2017 Jul 6. Plant Cell. 2017. PMID: 28684426 Free PMC article.
-
Histone variant H3.3 and its functions in reprogramming.Yi Chuan. 2018 Mar 20;40(3):186-196. doi: 10.16288/j.yczz.17-233. Yi Chuan. 2018. PMID: 29576542 Review.
-
All roads lead to chromatin: multiple pathways for histone deposition.Biochim Biophys Acta. 2013 Mar-Apr;1819(3-4):238-46. Biochim Biophys Acta. 2013. PMID: 24459726 Review.
Cited by
-
Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome.PLoS Genet. 2012;8(5):e1002658. doi: 10.1371/journal.pgen.1002658. Epub 2012 May 3. PLoS Genet. 2012. PMID: 22570629 Free PMC article.
-
The histone variant Sl_H2A.Z regulates carotenoid biosynthesis and gene expression during tomato fruit ripening.Hortic Res. 2021 Apr 1;8(1):85. doi: 10.1038/s41438-021-00520-3. Hortic Res. 2021. PMID: 33790255 Free PMC article.
-
Complementation of HYPONASTIC LEAVES1 by double-strand RNA-binding domains of DICER-LIKE1 in nuclear dicing bodies.Plant Physiol. 2013 Sep;163(1):108-17. doi: 10.1104/pp.113.219071. Epub 2013 Jul 25. Plant Physiol. 2013. PMID: 23886622 Free PMC article.
-
Nucleolus and chromatin.Histochem Cell Biol. 2018 Sep;150(3):209-225. doi: 10.1007/s00418-018-1696-3. Epub 2018 Jul 25. Histochem Cell Biol. 2018. PMID: 30046888 Free PMC article. Review.
-
The plant-specific histone residue Phe41 is important for genome-wide H3.1 distribution.Nat Commun. 2018 Feb 12;9(1):630. doi: 10.1038/s41467-018-02976-9. Nat Commun. 2018. PMID: 29434220 Free PMC article.
References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
-
- Talbert PB, Henikoff S. Histone variants—ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

