Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms
- PMID: 21670257
- PMCID: PMC3127913
- DOI: 10.1073/pnas.1106971108
Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms
Abstract
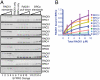
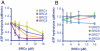
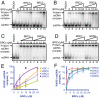
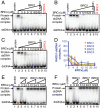
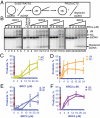
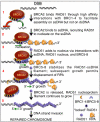
The human tumor suppressor protein BRCA2 plays a key role in recombinational DNA repair. BRCA2 recruits RAD51 to sites of DNA damage through interaction with eight conserved motifs of approximately 35 amino acids, the BRC repeats; however, the specific function of each repeat remains unclear. Here, we investigated the function of the individual BRC repeats by systematically analyzing their effects on RAD51 activities. Our results reveal the existence of two categories of BRC repeats that display unique functional characteristics. One group, comprising BRC1, -2, -3, and -4, binds to free RAD51 with high affinity. The second group, comprising BRC5, -6, -7, and -8, binds to free RAD51 with low affinity but binds to the RAD51-ssDNA filament with high affinity. Each member of the first group reduces the ATPase activity of RAD51, whereas none of the BRC repeats of the second group affects this activity. Thus, through different mechanisms, both types of BRC repeats bind to and stabilize the RAD51 nucleoprotein filament on ssDNA. In addition, members of the first group limit binding of RAD51 to duplex DNA, where members of the second group do not. Only the first group enhances DNA strand exchange by RAD51. Our results suggest that the two groups of BRC repeats have differentially evolved to ensure efficient formation of a nascent RAD51 filament on ssDNA by promoting its nucleation and growth, respectively. We propose that the BRC repeats cooperate in a partially redundant but reinforcing manner to ensure a high probability of RAD51 filament formation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
BRCA2 regulates DMC1-mediated recombination through the BRC repeats.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):3515-20. doi: 10.1073/pnas.1601691113. Epub 2016 Mar 14. Proc Natl Acad Sci U S A. 2016. PMID: 26976601 Free PMC article.
-
The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51.Cell. 2009 Mar 20;136(6):1032-43. doi: 10.1016/j.cell.2009.02.019. Cell. 2009. PMID: 19303847 Free PMC article.
-
A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange.Nucleic Acids Res. 2006;34(14):4000-11. doi: 10.1093/nar/gkl505. Epub 2006 Aug 16. Nucleic Acids Res. 2006. PMID: 16914443 Free PMC article.
-
Single-molecule imaging brings Rad51 nucleoprotein filaments into focus.Trends Cell Biol. 2010 May;20(5):269-76. doi: 10.1016/j.tcb.2010.02.004. Epub 2010 Mar 17. Trends Cell Biol. 2010. PMID: 20299221 Free PMC article. Review.
-
Unraveling the mechanism of BRCA2 in homologous recombination.Nat Struct Mol Biol. 2011 Jul 6;18(7):748-54. doi: 10.1038/nsmb.2096. Nat Struct Mol Biol. 2011. PMID: 21731065 Free PMC article. Review.
Cited by
-
Homologous recombination and its regulation.Nucleic Acids Res. 2012 Jul;40(13):5795-818. doi: 10.1093/nar/gks270. Epub 2012 Mar 30. Nucleic Acids Res. 2012. PMID: 22467216 Free PMC article. Review.
-
The phospho-dependent role of BRCA2 on the maintenance of chromosome integrity.Cell Cycle. 2021 Apr;20(8):731-741. doi: 10.1080/15384101.2021.1892994. Epub 2021 Mar 10. Cell Cycle. 2021. PMID: 33691600 Free PMC article. Review.
-
A Functional Analysis of the Unclassified Pro2767Ser BRCA2 Variant Reveals Its Potential Pathogenicity that Acts by Hampering DNA Binding and Homology-Mediated DNA Repair.Cancers (Basel). 2019 Sep 28;11(10):1454. doi: 10.3390/cancers11101454. Cancers (Basel). 2019. PMID: 31569370 Free PMC article.
-
Homologous recombination defects and how they affect replication fork maintenance.AIMS Genet. 2019 Apr 3;5(4):192-211. doi: 10.3934/genet.2018.4.192. eCollection 2018. AIMS Genet. 2019. PMID: 31435521 Free PMC article. Review.
-
Genome annotation by shotgun inactivation of a native gene in hemizygous cells: application to BRCA2 with implication of hypomorphic variants.Hum Mutat. 2015 Feb;36(2):260-9. doi: 10.1002/humu.22736. Hum Mutat. 2015. PMID: 25451944 Free PMC article.
References
-
- Lancaster JM, et al. BRCA2 mutations in primary breast and ovarian cancers. Nat Genet. 1996;13:238–240. - PubMed
-
- Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. - PubMed
-
- Sharan SK, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. - PubMed
-
- Yuan SS, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

