Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly
- PMID: 21670217
- PMCID: PMC3115798
- DOI: 10.1083/jcb.201103132
Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly
Erratum in
- J Cell Biol. 2011 Jul 11;194(1):155
- J Cell Biol. 2011 Jun 13;193(6):1101
Abstract
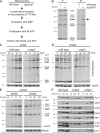
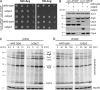
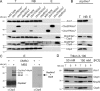
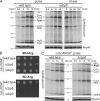
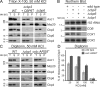
Mitochondria contain their own genetic system to express a small number of hydrophobic polypeptides, including cytochrome b, an essential subunit of the bc(1) complex of the respiratory chain. In this paper, we show in yeast that Cbp3, a bc(1) complex assembly factor, and Cbp6, a regulator of cytochrome b translation, form a complex that associates with the polypeptide tunnel exit of mitochondrial ribosomes and that exhibits two important functions in the biogenesis of cytochrome b. On the one hand, the interaction of Cbp3 and Cbp6 with mitochondrial ribosomes is necessary for efficient translation of cytochrome b transcript [corrected]. On the other hand, the Cbp3-Cbp6 complex interacts directly with newly synthesized cytochrome b in an assembly intermediate that is not ribosome bound and that contains the assembly factor Cbp4. Our results suggest that synthesis of cytochrome b occurs preferentially on those ribosomes that have the Cbp3-Cbp6 complex bound to their tunnel exit, an arrangement that may ensure tight coordination of cytochrome b synthesis and assembly.
Figures



Similar articles
-
Cbp3 and Cbp6 are dispensable for synthesis regulation of cytochrome b in yeast mitochondria.J Biol Chem. 2018 Apr 13;293(15):5585-5599. doi: 10.1074/jbc.RA117.000547. Epub 2018 Feb 23. J Biol Chem. 2018. PMID: 29475949 Free PMC article.
-
The Cbp3-Cbp6 complex coordinates cytochrome b synthesis with bc(1) complex assembly in yeast mitochondria.J Cell Biol. 2012 Oct 1;199(1):137-50. doi: 10.1083/jcb.201206040. Epub 2012 Sep 24. J Cell Biol. 2012. PMID: 23007649 Free PMC article.
-
Molecular Wiring of a Mitochondrial Translational Feedback Loop.Mol Cell. 2020 Feb 20;77(4):887-900.e5. doi: 10.1016/j.molcel.2019.11.019. Epub 2019 Dec 26. Mol Cell. 2020. PMID: 31883951
-
The polypeptide tunnel exit of the mitochondrial ribosome is tailored to meet the specific requirements of the organelle.Bioessays. 2010 Dec;32(12):1050-7. doi: 10.1002/bies.201000081. Epub 2010 Oct 21. Bioessays. 2010. PMID: 20967780 Review.
-
Activation of Yeast Mitochondrial Translation: Who Is in Charge?Biochemistry (Mosc). 2018 Feb;83(2):87-97. doi: 10.1134/S0006297918020013. Biochemistry (Mosc). 2018. PMID: 29618295 Review.
Cited by
-
Insertion Defects of Mitochondrially Encoded Proteins Burden the Mitochondrial Quality Control System.Cells. 2018 Oct 17;7(10):172. doi: 10.3390/cells7100172. Cells. 2018. PMID: 30336542 Free PMC article.
-
Structure of the yeast mitochondrial large ribosomal subunit.Science. 2014 Mar 28;343(6178):1485-1489. doi: 10.1126/science.1249410. Science. 2014. PMID: 24675956 Free PMC article.
-
Biogenesis of the cytochrome bc(1) complex and role of assembly factors.Biochim Biophys Acta. 2012 Feb;1817(2):276-86. doi: 10.1016/j.bbabio.2011.11.009. Epub 2011 Nov 22. Biochim Biophys Acta. 2012. Corrected and republished in: Biochim Biophys Acta. 2012 Jun;1817(6):872-82. doi: 10.1016/j.bbabio.2012.03.003 PMID: 22138626 Free PMC article. Corrected and republished. Review.
-
Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.EMBO Rep. 2020 Dec 3;21(12):e51015. doi: 10.15252/embr.202051015. Epub 2020 Oct 5. EMBO Rep. 2020. PMID: 33016568 Free PMC article.
-
Cbp3 and Cbp6 are dispensable for synthesis regulation of cytochrome b in yeast mitochondria.J Biol Chem. 2018 Apr 13;293(15):5585-5599. doi: 10.1074/jbc.RA117.000547. Epub 2018 Feb 23. J Biol Chem. 2018. PMID: 29475949 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

