Identification and functional analysis of 9p24 amplified genes in human breast cancer
- PMID: 21666724
- PMCID: PMC3886828
- DOI: 10.1038/onc.2011.227
Identification and functional analysis of 9p24 amplified genes in human breast cancer
Abstract
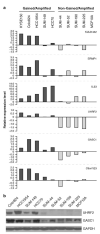
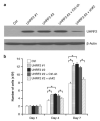
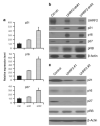
Previously, our group identified a novel amplicon at chromosome 9p24 in human esophageal and breast cancers, and cloned the novel gene, GASC1 (gene amplified in squamous cell carcinoma 1, also known as JMJD2C/KDM4C), from this amplicon. GASC1 is a histone demethylase involved in the deregulation of histone methylation in cancer cells. In the current study, we aimed to comprehensively characterize the genes in the 9p24 amplicon in human breast cancer. We performed extensive genomic analyses on a panel of cancer cell lines and narrowed the shortest region of overlap to approximately 2 Mb. Based on statistical analysis of copy number increase and overexpression, the 9p24 amplicon contains six candidate oncogenes. Among these, four genes (GASC1 UHRF2, KIAA1432 and C9orf123) are overexpressed only in the context of gene amplification while two genes (ERMP1 and IL33) are overexpressed independent of the copy number increase. We then focused our studies on the UHRF2 gene, which has a potential involvement in both DNA methylation and histone modification. Knocking down UHRF2 expression inhibited the growth of breast cancer cells specifically with 9p24 amplification. Conversely, ectopic overexpression of UHRF2 in non-tumorigenic MCF10A cells promoted cell proliferation. Furthermore, we demonstrated that UHRF2 has the ability to suppress the expression of key cell-cycle inhibitors, such as p16(INK4a), p21(Waf1/Cip1) and p27(Kip1). Taken together, our studies support the notion that the 9p24 amplicon contains multiple oncogenes that may integrate genetic and epigenetic codes and have important roles in human tumorigenesis.
Figures


Similar articles
-
Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer.Oncogene. 2009 Dec 17;28(50):4491-500. doi: 10.1038/onc.2009.297. Epub 2009 Sep 28. Oncogene. 2009. PMID: 19784073 Free PMC article.
-
Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines.Cancer Res. 2000 Sep 1;60(17):4735-9. Cancer Res. 2000. PMID: 10987278
-
Cortactin modulates RhoA activation and expression of Cip/Kip cyclin-dependent kinase inhibitors to promote cell cycle progression in 11q13-amplified head and neck squamous cell carcinoma cells.Mol Cell Biol. 2010 Nov;30(21):5057-70. doi: 10.1128/MCB.00249-10. Epub 2010 Aug 30. Mol Cell Biol. 2010. PMID: 20805359 Free PMC article.
-
A novel amplicon at 9p23 - 24 in squamous cell carcinoma of the esophagus that lies proximal to GASC1 and harbors NFIB.Jpn J Cancer Res. 2001 Apr;92(4):423-8. doi: 10.1111/j.1349-7006.2001.tb01112.x. Jpn J Cancer Res. 2001. PMID: 11346465 Free PMC article.
-
The 17q23 amplicon and breast cancer.Breast Cancer Res Treat. 2003 Apr;78(3):313-22. doi: 10.1023/a:1023081624133. Breast Cancer Res Treat. 2003. PMID: 12755490 Review.
Cited by
-
Multidimensional Proteomics Reveals a Role of UHRF2 in the Regulation of Epithelial-Mesenchymal Transition (EMT).Mol Cell Proteomics. 2016 Jul;15(7):2263-78. doi: 10.1074/mcp.M115.057448. Epub 2016 Apr 25. Mol Cell Proteomics. 2016. PMID: 27114453 Free PMC article.
-
miR-148b Functions as a Tumor Suppressor by Targeting Endoplasmic Reticulum Metallo Protease 1 in Human Endometrial Cancer Cells.Oncol Res. 2018 Dec 27;27(1):81-88. doi: 10.3727/096504018X15202988139874. Epub 2018 Mar 9. Oncol Res. 2018. Retraction in: Oncol Res. 2024 Jul 17;32(8):1379. doi: 10.32604/or.2024.055034. PMID: 29523216 Free PMC article. Retracted.
-
A community effort to assess and improve drug sensitivity prediction algorithms.Nat Biotechnol. 2014 Dec;32(12):1202-12. doi: 10.1038/nbt.2877. Epub 2014 Jun 1. Nat Biotechnol. 2014. PMID: 24880487 Free PMC article.
-
Histone demethylase JMJD2C: epigenetic regulators in tumors.Oncotarget. 2017 Jul 12;8(53):91723-91733. doi: 10.18632/oncotarget.19176. eCollection 2017 Oct 31. Oncotarget. 2017. PMID: 29207681 Free PMC article. Review.
-
Kdm4c is Recruited to Mitotic Chromosomes and Is Relevant for Chromosomal Stability, Cell Migration and Invasion of Triple Negative Breast Cancer Cells.Breast Cancer (Auckl). 2018 Jul 27;12:1178223418773075. doi: 10.1177/1178223418773075. eCollection 2018. Breast Cancer (Auckl). 2018. PMID: 30083054 Free PMC article.
References
-
- Akiyama M, Ishida N, Ogawa T, Yogo K, Takeya T. Molecular cloning and functional analysis of a novel Cx43 partner protein CIP150. Biochem Biophys Res Commun. 2005;335:1264–1271. - PubMed
-
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. - PubMed
-
- Albertson DG, Collins C, McCormick F, Gray JW. Chromo-some aberrations in solid tumors. Nat Genet. 2003;34:369–376. - PubMed
-
- Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther. 2007;115:419–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

