Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity
- PMID: 21653673
- PMCID: PMC3147955
- DOI: 10.1128/JVI.00756-11
Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity
Abstract
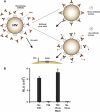
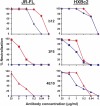
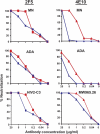
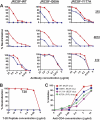
On the prereceptor-engaged HIV-1 envelope glycoprotein (Env) spike, epitope access by the membrane-proximal external region (MPER)-directed broadly neutralizing antibodies 2F5 and 4E10 remains unresolved. Data on binding to cell surface Env and entry data using primary isolates suggest inaccessibility of the 2F5 and 4E10 epitopes on the viral spike prior to receptor engagement, but trimer gel shift analysis and slow kinetics of shedding induced by 2F5 and 4E10 indicate otherwise. Therefore, it remains unclear if the epitopes themselves are formed in their antibody-bound state (or at least sampled) prior to receptor/coreceptor engagement or if receptor interactions both expose and form the MPER epitopes, presumably in the putative prefusion transitional intermediate. Here, we performed antibody-virus "washout experiments" using both lab-adapted and a panel of clade B primary isolates to analyze MPER accessibility. The neutralization activity of 2F5 and 4E10 against lab-adapted viruses and sensitive and moderately resistant viruses was largely unaffected by relatively rapid antibody-virus washing, suggesting direct interaction with the "static" spike. However, for more neutralization-resistant viruses, the 2F5 and 4E10 antibodies could neutralize only under the "no antibody-virus wash" conditions, implying that the MPER epitopes were not accessible prior to receptor engagement. Accessibility in the washout conditions could be precisely predicted by the relative resistance to neutralization in a standard neutralization format. These data are consistent with a model in which the local MPER antibody epitope conformations may be sampled on the native spike but are occluded to antibody by local steric or distal quaternary constraints adopted by highly resistant HIV-1 isolates.
Figures


Similar articles
-
HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies.AIDS Res Hum Retroviruses. 2011 Aug;27(8):877-87. doi: 10.1089/AID.2010.0281. Epub 2011 Jan 19. AIDS Res Hum Retroviruses. 2011. PMID: 21158699 Free PMC article.
-
An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10.J Virol. 2007 Apr;81(8):4033-43. doi: 10.1128/JVI.02588-06. Epub 2007 Feb 7. J Virol. 2007. PMID: 17287272 Free PMC article.
-
Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1.J Virol. 2005 Jan;79(2):1252-61. doi: 10.1128/JVI.79.2.1252-1261.2005. J Virol. 2005. PMID: 15613352 Free PMC article.
-
The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design.Microbiol Mol Biol Rev. 2008 Mar;72(1):54-84, table of contents. doi: 10.1128/MMBR.00020-07. Microbiol Mol Biol Rev. 2008. PMID: 18322034 Free PMC article. Review.
-
Neutralizing Antibodies Targeting HIV-1 gp41.Viruses. 2020 Oct 23;12(11):1210. doi: 10.3390/v12111210. Viruses. 2020. PMID: 33114242 Free PMC article. Review.
Cited by
-
Structural basis for broad detection of genogroup II noroviruses by a monoclonal antibody that binds to a site occluded in the viral particle.J Virol. 2012 Apr;86(7):3635-46. doi: 10.1128/JVI.06868-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278249 Free PMC article.
-
Membrane HIV-1 envelope glycoproteins stabilized more strongly in a pretriggered conformation than natural virus Envs.iScience. 2024 May 28;27(7):110141. doi: 10.1016/j.isci.2024.110141. eCollection 2024 Jul 19. iScience. 2024. PMID: 38979012 Free PMC article.
-
Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.Cell Host Microbe. 2019 Nov 13;26(5):623-637.e8. doi: 10.1016/j.chom.2019.09.016. Epub 2019 Oct 22. Cell Host Microbe. 2019. PMID: 31653484 Free PMC article.
-
Neutralizing IgG at the portal of infection mediates protection against vaginal simian/human immunodeficiency virus challenge.J Virol. 2013 Nov;87(21):11604-16. doi: 10.1128/JVI.01361-13. Epub 2013 Aug 21. J Virol. 2013. PMID: 23966410 Free PMC article.
-
Antibody to gp41 MPER alters functional properties of HIV-1 Env without complete neutralization.PLoS Pathog. 2014 Jul 24;10(7):e1004271. doi: 10.1371/journal.ppat.1004271. eCollection 2014 Jul. PLoS Pathog. 2014. PMID: 25058619 Free PMC article.
References
-
- Burton D. R., et al. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233–236 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

