A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells
- PMID: 21653320
- PMCID: PMC3152493
- DOI: 10.1182/blood-2011-02-337360
A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells
Abstract
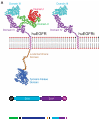
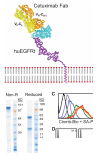
An unmet need in cell engineering is the availability of a single transgene encoded, functionally inert, human polypeptide that can serve multiple purposes, including ex vivo cell selection, in vivo cell tracking, and as a target for in vivo cell ablation. Here we describe a truncated human EGFR polypeptide (huEGFRt) that is devoid of extracellular N-terminal ligand binding domains and intracellular receptor tyrosine kinase activity but retains the native amino acid sequence, type I transmembrane cell surface localization, and a conformationally intact binding epitope for pharmaceutical-grade anti-EGFR monoclonal antibody, cetuximab (Erbitux). After lentiviral transduction of human T cells with vectors that coordinately express tumor-specific chimeric antigen receptors and huEGFRt, we show that huEGFRt serves as a highly efficient selection epitope for chimeric antigen receptor(+) T cells using biotinylated cetuximab in conjunction with current good manufacturing practices (cGMP)-grade anti-biotin immunomagnetic microbeads. Moreover, huEGFRt provides a cell surface marker for in vivo tracking of adoptively transferred T cells using both flow cytometry and immunohistochemistry, and a target for cetuximab-mediated antibody-dependent cellular cytotoxicity and in vivo elimination. The versatility of huEGFRt and the availability of pharmaceutical-grade reagents for its clinical application denote huEGFRt as a significant new tool for cellular engineering.
Figures


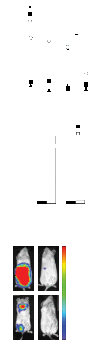
Similar articles
-
A Cetuximab-Mediated Suicide System in Chimeric Antigen Receptor-Modified Hematopoietic Stem Cells for Cancer Therapy.Hum Gene Ther. 2019 Apr;30(4):413-428. doi: 10.1089/hum.2018.180. Hum Gene Ther. 2019. PMID: 30860401 Free PMC article.
-
Antibody with Infinite Affinity for In Vivo Tracking of Genetically Engineered Lymphocytes.J Nucl Med. 2018 Dec;59(12):1894-1900. doi: 10.2967/jnumed.118.208041. Epub 2018 Jun 14. J Nucl Med. 2018. PMID: 29903928 Free PMC article.
-
A transgene-encoded truncated human epidermal growth factor receptor for depletion of anti- B-cell maturation antigen CAR-T cells.Cell Immunol. 2021 May;363:104342. doi: 10.1016/j.cellimm.2021.104342. Epub 2021 Mar 14. Cell Immunol. 2021. PMID: 33765541
-
Optimization of Human NK Cell Manufacturing: Fully Automated Separation, Improved Ex Vivo Expansion Using IL-21 with Autologous Feeder Cells, and Generation of Anti-CD123-CAR-Expressing Effector Cells.Hum Gene Ther. 2017 Oct;28(10):897-913. doi: 10.1089/hum.2017.157. Epub 2017 Aug 15. Hum Gene Ther. 2017. PMID: 28810809
-
Cetuximab: an epidermal growth factor receptor chemeric human-murine monoclonal antibody.Drugs Today (Barc). 2005 Feb;41(2):107-27. doi: 10.1358/dot.2005.41.2.882662. Drugs Today (Barc). 2005. PMID: 15821783 Review.
Cited by
-
Promises and Pitfalls of Next-Generation Treg Adoptive Immunotherapy.Cancers (Basel). 2023 Dec 17;15(24):5877. doi: 10.3390/cancers15245877. Cancers (Basel). 2023. PMID: 38136421 Free PMC article. Review.
-
IL-12 secreting tumor-targeted chimeric antigen receptor T cells eradicate ovarian tumors in vivo.Oncoimmunology. 2015 Jan 23;4(3):e994446. doi: 10.4161/2162402X.2014.994446. eCollection 2015 Mar. Oncoimmunology. 2015. PMID: 25949921 Free PMC article.
-
Engineering Strategies to Enhance TCR-Based Adoptive T Cell Therapy.Cells. 2020 Jun 18;9(6):1485. doi: 10.3390/cells9061485. Cells. 2020. PMID: 32570906 Free PMC article. Review.
-
Promoter usage regulating the surface density of CAR molecules may modulate the kinetics of CAR-T cells in vivo.Mol Ther Methods Clin Dev. 2021 Mar 13;21:237-246. doi: 10.1016/j.omtm.2021.03.007. eCollection 2021 Jun 11. Mol Ther Methods Clin Dev. 2021. PMID: 33869653 Free PMC article.
-
Safety switch optimization enhances antibody-mediated elimination of CAR T cells.Front Mol Med. 2022 Oct 11;2:1026474. doi: 10.3389/fmmed.2022.1026474. eCollection 2022. Front Mol Med. 2022. PMID: 39086975 Free PMC article.
References
-
- Cooper LJ, Kalos M, DiGiusto D, et al. T-cell genetic modification for re-directed tumor recognition. Cancer Chemother Biol Response Modif. 2005;22:293–324. - PubMed
-
- Fehse B, Richters A, Putimtseva-Scharf K, et al. CD34 splice variant: an attractive marker for selection of gene-modified cells. Mol Ther. 2000;1(5):448–456. - PubMed
-
- Fecci PE, Ochiai H, Mitchell DA, et al. Systemic CTLA-4 blockade ameliorates glioma-induced changes to the CD4+ T cell compartment without affecting regulatory T-cell function. Clin Cancer Res. 2007;13(7):2158–2167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

