A multifunctional bioconjugate module for versatile photoaffinity labeling and click chemistry of RNA
- PMID: 21646334
- PMCID: PMC3167637
- DOI: 10.1093/nar/gkr449
A multifunctional bioconjugate module for versatile photoaffinity labeling and click chemistry of RNA
Abstract
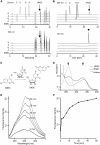
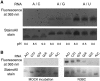
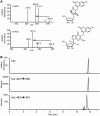
A multifunctional reagent based on a coumarin scaffold was developed for derivatization of naive RNA. The alkylating agent N3BC [7-azido-4-(bromomethyl)coumarin], obtained by Pechmann condensation, is selective for uridine. N3BC and its RNA conjugates are pre-fluorophores which permits controlled modular and stepwise RNA derivatization. The success of RNA alkylation by N3BC can be monitored by photolysis of the azido moiety, which generates a coumarin fluorophore that can be excited with UV light of 320 nm. The azidocoumarin-modified RNA can be flexibly employed in structure-function studies. Versatile applications include direct use in photo-crosslinking studies to cognate proteins, as demonstrated with tRNA and RNA fragments from the MS2 phage and the HIV genome. Alternatively, the azide function can be used for further derivatization by click-chemistry. This allows e.g. the introduction of an additional fluorophore for excitation with visible light.
Figures
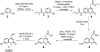



Similar articles
-
Clickable photoaffinity ligands for the human serotonin transporter based on the selective serotonin reuptake inhibitor (S)-citalopram.Bioorg Med Chem Lett. 2018 Nov 15;28(21):3431-3435. doi: 10.1016/j.bmcl.2018.09.029. Epub 2018 Sep 22. Bioorg Med Chem Lett. 2018. PMID: 30266542 Free PMC article.
-
Tyrosine specific sequential labeling of proteins.Bioorg Med Chem Lett. 2013 Nov 1;23(21):5776-8. doi: 10.1016/j.bmcl.2013.09.002. Epub 2013 Sep 8. Bioorg Med Chem Lett. 2013. PMID: 24075730
-
Development of surface immobilized 3-azidocoumarin-based fluorogenic probe via strain promoted click chemistry.Bioorg Med Chem Lett. 2015 Dec 15;25(24):5737-42. doi: 10.1016/j.bmcl.2015.10.078. Epub 2015 Oct 26. Bioorg Med Chem Lett. 2015. PMID: 26531149
-
[Molecular Conjugation for the "Diazido Probe" Method].Yakugaku Zasshi. 2018;138(8):1049-1058. doi: 10.1248/yakushi.17-00211-4. Yakugaku Zasshi. 2018. PMID: 30068846 Review. Japanese.
-
Current approaches for RNA labeling in vitro and in cells based on click reactions.Chembiochem. 2014 Nov 3;15(16):2342-7. doi: 10.1002/cbic.201402240. Epub 2014 Sep 15. Chembiochem. 2014. PMID: 25224574 Review.
Cited by
-
Co-mapping studies of QTLs for fruit acidity and candidate genes of organic acid metabolism and proton transport in sweet melon (Cucumis melo L.).Theor Appl Genet. 2012 Jul;125(2):343-53. doi: 10.1007/s00122-012-1837-3. Epub 2012 Mar 10. Theor Appl Genet. 2012. PMID: 22406955
-
Remarkable acceleration of a DNA/RNA inter-strand functionality transfer reaction to modify a cytosine residue: the proximity effect via complexation with a metal cation.Nucleic Acids Res. 2014 Jul;42(13):8808-15. doi: 10.1093/nar/gku538. Epub 2014 Jun 23. Nucleic Acids Res. 2014. PMID: 24957600 Free PMC article.
-
Spatially Mediated Paper Reactors for On-Site Multicoded Encryption.JACS Au. 2024 Apr 22;4(6):2151-2159. doi: 10.1021/jacsau.4c00062. eCollection 2024 Jun 24. JACS Au. 2024. PMID: 38938820 Free PMC article.
-
Ribonucleic Acid Sequence Characterization by Negative Electron Transfer Dissociation Mass Spectrometry.Anal Chem. 2020 Mar 17;92(6):4436-4444. doi: 10.1021/acs.analchem.9b05388. Epub 2020 Mar 5. Anal Chem. 2020. PMID: 32091202 Free PMC article.
-
5'-deoxy-5'-hydrazinylguanosine as an initiator of T7 Rna polymerase-catalyzed transcriptions for the preparation of labeling-ready RNAs.Nucleosides Nucleotides Nucleic Acids. 2013;32(12):670-81. doi: 10.1080/15257770.2013.851393. Nucleosides Nucleotides Nucleic Acids. 2013. PMID: 24328564 Free PMC article.
References
-
- Gait MJ. DNA/RNA synthesis and labelling. Curr. Opin. Biotechnol. 1991;2:61–68. - PubMed
-
- Sproat BS. Chemistry and applications of oligonucleotide analogues. J. Biotechnol. 1995;41:221–238. - PubMed
-
- Verma S, Eckstein F. Modified oligonucleotides: synthesis and strategy for users. Annu. Rev. Biochem. 1998;67:99–134. - PubMed
-
- Cochrane JC, Strobel SA. Probing RNA structure and function by nucleotide analog interference mapping. Curr. Protoc. Nucleic Acid. Chem. 2004 Chapter 6, Unit 6.9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

