Mutational analysis of three predicted 5'-proximal stem-loop structures in the genome of tick-borne encephalitis virus indicates different roles in RNA replication and translation
- PMID: 21645915
- PMCID: PMC3182534
- DOI: 10.1016/j.virol.2011.05.008
Mutational analysis of three predicted 5'-proximal stem-loop structures in the genome of tick-borne encephalitis virus indicates different roles in RNA replication and translation
Abstract
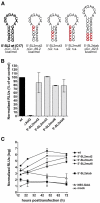
Flavivirus gene expression is modulated by RNA secondary structure elements at the terminal ends of the viral RNA molecule. For tick-borne encephalitis virus (TBEV), four stem-loop (SL) elements have been predicted in the first 180 nucleotides of the viral genome: 5'-SL1, 5'-SL2, 5'-SL3 and 5'-SL4. The last three of these appear to be unique to tick-borne flaviviruses. Here, we report their characterization by mutagenesis in a TBEV luciferase reporter system. By manipulating their thermodynamic properties, we found that an optimal stability of the 5'-SL2 is required for efficient RNA replication. 5'-SL3 formation is also important for viral RNA replication, but although it contains the viral start codon, its formation is dispensable for RNA translation. 5'-SL4 appears to facilitate both RNA translation and replication. Our data suggest that maintenance of the balanced thermodynamic stability of these SL elements is important for temporal regulation of its different functions.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Model System for the Formation of Tick-Borne Encephalitis Virus Replication Compartments without Viral RNA Replication.J Virol. 2019 Aug 28;93(18):e00292-19. doi: 10.1128/JVI.00292-19. Print 2019 Sep 15. J Virol. 2019. PMID: 31243132 Free PMC article.
-
Functional characterization of 5' untranslated region (UTR) secondary RNA structures in the replication of tick-borne encephalitis virus in mammalian cells.PLoS Negl Trop Dis. 2023 Jan 23;17(1):e0011098. doi: 10.1371/journal.pntd.0011098. eCollection 2023 Jan. PLoS Negl Trop Dis. 2023. PMID: 36689554 Free PMC article.
-
The stress granule component TIA-1 binds tick-borne encephalitis virus RNA and is recruited to perinuclear sites of viral replication to inhibit viral translation.J Virol. 2014 Jun;88(12):6611-22. doi: 10.1128/JVI.03736-13. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696465 Free PMC article.
-
Visual detection of Flavivirus RNA in living cells.Methods. 2016 Apr 1;98:82-90. doi: 10.1016/j.ymeth.2015.11.002. Epub 2015 Nov 2. Methods. 2016. PMID: 26542763 Free PMC article. Review.
-
Steps of the tick-borne encephalitis virus replication cycle that affect neuropathogenesis.Virus Res. 2005 Aug;111(2):161-74. doi: 10.1016/j.virusres.2005.04.007. Virus Res. 2005. PMID: 15871909 Review.
Cited by
-
Molecular Mechanisms of Interaction Between Human Immune Cells and Far Eastern Tick-Borne Encephalitis Virus Strains.Viral Immunol. 2015 Jun;28(5):272-81. doi: 10.1089/vim.2014.0083. Epub 2015 Feb 19. Viral Immunol. 2015. PMID: 25695407 Free PMC article.
-
A novel coding-region RNA element modulates infectious dengue virus particle production in both mammalian and mosquito cells and regulates viral replication in Aedes aegypti mosquitoes.Virology. 2012 Oct 25;432(2):511-26. doi: 10.1016/j.virol.2012.06.028. Epub 2012 Jul 25. Virology. 2012. PMID: 22840606 Free PMC article.
-
The 5' and 3' Untranslated Regions of the Flaviviral Genome.Viruses. 2017 Jun 6;9(6):137. doi: 10.3390/v9060137. Viruses. 2017. PMID: 28587300 Free PMC article. Review.
-
Functional Information Stored in the Conserved Structural RNA Domains of Flavivirus Genomes.Front Microbiol. 2017 Apr 3;8:546. doi: 10.3389/fmicb.2017.00546. eCollection 2017. Front Microbiol. 2017. PMID: 28421048 Free PMC article. Review.
-
Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization.Elife. 2016 Oct 1;5:e17636. doi: 10.7554/eLife.17636. Elife. 2016. PMID: 27692070 Free PMC article.
References
-
- Alvarez DE, Filomatori CV, Gamarnik AV. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology. 2008;375(1):223–235. - PubMed
-
- Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162(2):290–299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

