Transcription factors Sp1 and Sp4 regulate TRPV1 gene expression in rat sensory neurons
- PMID: 21645329
- PMCID: PMC3121596
- DOI: 10.1186/1744-8069-7-44
Transcription factors Sp1 and Sp4 regulate TRPV1 gene expression in rat sensory neurons
Abstract
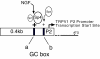
Background: The capsaicin receptor, transient receptor potential vanilloid type -1 (TRPV1) directs complex roles in signal transduction including the detection of noxious stimuli arising from cellular injury and inflammation. Under pathophysiologic conditions, TRPV1 mRNA and receptor protein expression are elevated in dorsal root ganglion (DRG) neurons for weeks to months and is associated with hyperalgesia. Building on our previous isolation of a promoter system for the rat TRPV1 gene, we investigated the proximal TRPV1 P2-promoter by first identifying candidate Sp1-like transcription factors bound in vivo to the P2-promoter using chromatin immunoprecipitation (ChIP) assay. We then performed deletion analysis of GC-box binding sites, and quantified promoter activity under conditions of Sp1 / Sp4 over-expression versus inhibition/knockdown. mRNA encoding Sp1, Sp4 and TRPV1 were quantified by qRT-PCR under conditions of Sp1/Sp4 over-expression or siRNA mediated knockdown in cultured DRG neurons.
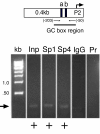
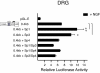
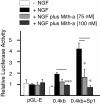
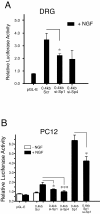
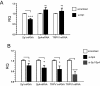
Results: Using ChIP analysis of DRG tissue, we demonstrated that Sp1 and Sp4 are bound to the candidate GC-box site region within the endogenous TRPV1 P2-promoter. Deletion of GC-box "a" or "a + b" within the P2- promoter resulted in a complete loss of transcriptional activity indicating that GC-box "a" was the critical site for promoter activation. Co-transfection of Sp1 increased P2-promoter activity in cultured DRG neurons whereas mithramycin-a, an inhibitor of Sp1-like function, dose dependently blocked NGF and Sp1-dependent promoter activity in PC12 cells. Co-transfection of siRNA directed against Sp1 or Sp4 decreased promoter activity in DRG neurons and NGF treated PC12 cells. Finally, electroporation of Sp1 or Sp4 cDNA into cultures of DRG neurons directed an increase in Sp1/Sp4 mRNA and importantly an increase in TRPV1 mRNA. Conversely, combined si-RNA directed knockdown of Sp1/Sp4 resulted in a decrease in TRPV1 mRNA.
Conclusion: Based on these studies, we now propose a model of TRPV1 expression that is dependent on Sp1-like transcription factors with Sp4 playing a predominant role in activating TRPV1 RNA transcription in DRG neurons. Given that increases of TRPV1 expression have been implicated in a wide range of pathophysiologic states including persistent painful conditions, blockade of Sp1-like transcription factors represents a novel direction in therapeutic strategies.
Figures



Similar articles
-
The anticancer antibiotic mithramycin-A inhibits TRPV1 expression in dorsal root ganglion neurons.Neurosci Lett. 2014 Aug 22;578:211-6. doi: 10.1016/j.neulet.2014.01.021. Epub 2014 Jan 25. Neurosci Lett. 2014. PMID: 24468003
-
Transcription of rat TRPV1 utilizes a dual promoter system that is positively regulated by nerve growth factor.J Neurochem. 2007 Apr;101(1):212-22. doi: 10.1111/j.1471-4159.2006.04363.x. Epub 2007 Jan 8. J Neurochem. 2007. PMID: 17217411
-
Transcription factor Sp4 is required for hyperalgesic state persistence.PLoS One. 2019 Feb 27;14(2):e0211349. doi: 10.1371/journal.pone.0211349. eCollection 2019. PLoS One. 2019. PMID: 30811405 Free PMC article.
-
Specificity Proteins (Sp) and Cancer.Int J Mol Sci. 2023 Mar 8;24(6):5164. doi: 10.3390/ijms24065164. Int J Mol Sci. 2023. PMID: 36982239 Free PMC article. Review.
-
TRPV1 splice variants: structure and function.Front Biosci (Landmark Ed). 2010 Jun 1;15(3):872-82. doi: 10.2741/3651. Front Biosci (Landmark Ed). 2010. PMID: 20515731 Free PMC article. Review.
Cited by
-
Impact of diabetes mellitus on bladder uroepithelial cells.Am J Physiol Regul Integr Comp Physiol. 2013 Jan 15;304(2):R84-93. doi: 10.1152/ajpregu.00129.2012. Epub 2012 Nov 21. Am J Physiol Regul Integr Comp Physiol. 2013. PMID: 23174855 Free PMC article.
-
Chemotherapy for pain: reversing inflammatory and neuropathic pain with the anticancer agent mithramycin A.Pain. 2024 Jan 1;165(1):54-74. doi: 10.1097/j.pain.0000000000002972. Epub 2023 Jun 27. Pain. 2024. PMID: 37366593 Free PMC article.
-
Capsaicin, Nociception and Pain.Molecules. 2016 Jun 18;21(6):797. doi: 10.3390/molecules21060797. Molecules. 2016. PMID: 27322240 Free PMC article. Review.
-
The Mysteries of Capsaicin-Sensitive Afferents.Front Physiol. 2020 Dec 16;11:554195. doi: 10.3389/fphys.2020.554195. eCollection 2020. Front Physiol. 2020. PMID: 33391007 Free PMC article. Review.
-
A vicious cycle between acid sensing and survival signaling in myeloma cells: acid-induced epigenetic alteration.Oncotarget. 2016 Oct 25;7(43):70447-70461. doi: 10.18632/oncotarget.11927. Oncotarget. 2016. PMID: 27626482 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

