Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice
- PMID: 21628403
- PMCID: PMC3622520
- DOI: 10.1182/blood-2011-02-338848
Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice
Abstract
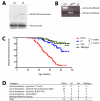
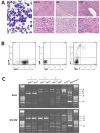
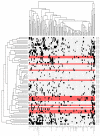
The t(12;21) translocation that generates the ETV6-RUNX1 (TEL-AML1) fusion gene, is the most common chromosomal rearrangement in childhood cancer and is exclusively associated with B-cell precursor acute lymphoblastic leukemia (BCP-ALL). The translocation arises in utero and is necessary but insufficient for the development of leukemia. Single-nucleotide polymorphism array analysis of ETV6-RUNX1 patient samples has identified multiple additional genetic alterations; however, the role of these lesions in leukemogenesis remains undetermined. Moreover, murine models of ETV6-RUNX1 ALL that faithfully recapitulate the human disease are lacking. To identify novel genes that cooperate with ETV6-RUNX1 in leukemogenesis, we generated a mouse model that uses the endogenous Etv6 locus to coexpress the Etv6-RUNX1 fusion and Sleeping Beauty transposase. An insertional mutagenesis screen was performed by intercrossing these mice with those carrying a Sleeping Beauty transposon array. In contrast to previous models, a substantial proportion (20%) of the offspring developed BCP-ALL. Isolation of the transposon insertion sites identified genes known to be associated with BCP-ALL, including Ebf1 and Epor, in addition to other novel candidates. This is the first mouse model of ETV6-RUNX1 to develop BCP-ALL and provides important insight into the cooperating genetic alterations in ETV6-RUNX1 leukemia.
Figures

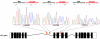
Similar articles
-
ETV6/RUNX1-like acute lymphoblastic leukemia: A novel B-cell precursor leukemia subtype associated with the CD27/CD44 immunophenotype.Genes Chromosomes Cancer. 2017 Aug;56(8):608-616. doi: 10.1002/gcc.22464. Epub 2017 May 5. Genes Chromosomes Cancer. 2017. PMID: 28395118
-
Prevalence of ETV6-RUNX1 fusion gene in children with acute lymphoblastic leukemia in China.Cancer Genet Cytogenet. 2007 Oct 1;178(1):57-60. doi: 10.1016/j.cancergencyto.2007.05.019. Cancer Genet Cytogenet. 2007. PMID: 17889709
-
Abnormalities of the der(12)t(12;21) in ETV6-RUNX1 acute lymphoblastic leukemia.Genes Chromosomes Cancer. 2013 Feb;52(2):202-13. doi: 10.1002/gcc.22021. Epub 2012 Oct 18. Genes Chromosomes Cancer. 2013. PMID: 23077088
-
Siblings with ETV6/RUNX1-positive B-lymphoblastic leukemia: A single site experience and review of the literature.Ann Diagn Pathol. 2020 Oct;48:151588. doi: 10.1016/j.anndiagpath.2020.151588. Epub 2020 Aug 14. Ann Diagn Pathol. 2020. PMID: 32836179 Review.
-
The Landscape of Secondary Genetic Rearrangements in Pediatric Patients with B-Cell Acute Lymphoblastic Leukemia with t(12;21).Cells. 2023 Jan 18;12(3):357. doi: 10.3390/cells12030357. Cells. 2023. PMID: 36766699 Free PMC article. Review.
Cited by
-
WNT inhibitory factor 1 (WIF1) is a novel fusion partner of RUNX family transcription factor 1 (RUNX1) in acute myeloid leukemia with t(12;21)(q14;q22).J Hematop. 2024 Dec;17(4):245-249. doi: 10.1007/s12308-024-00597-4. Epub 2024 Jul 27. J Hematop. 2024. PMID: 39066949
-
Cancer gene discovery: exploiting insertional mutagenesis.Mol Cancer Res. 2013 Oct;11(10):1141-58. doi: 10.1158/1541-7786.MCR-13-0244. Epub 2013 Aug 8. Mol Cancer Res. 2013. PMID: 23928056 Free PMC article. Review.
-
Transposon Insertion Mutagenesis in Mice for Modeling Human Cancers: Critical Insights Gained and New Opportunities.Int J Mol Sci. 2020 Feb 10;21(3):1172. doi: 10.3390/ijms21031172. Int J Mol Sci. 2020. PMID: 32050713 Free PMC article. Review.
-
Genome-wide interference of ZNF423 with B-lineage transcriptional circuitries in acute lymphoblastic leukemia.Blood Adv. 2021 Mar 9;5(5):1209-1223. doi: 10.1182/bloodadvances.2020001844. Blood Adv. 2021. PMID: 33646306 Free PMC article.
-
Initially disadvantaged, TEL-AML1 cells expand and initiate leukemia in response to irradiation and cooperating mutations.Leukemia. 2013 Jul;27(7):1570-3. doi: 10.1038/leu.2013.15. Epub 2013 Jan 16. Leukemia. 2013. PMID: 23443342 Free PMC article. No abstract available.
References
-
- Romana SP, Mauchauffé M, Le Coniat M, et al. The t(12;21) of acute lymphoblastic leukemia results in a TEL-AML1 gene fusion. Blood. 1995;85:3662–3670. - PubMed
-
- Shurtleff SA, Buijs A, Behm FG, et al. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. - PubMed
-
- Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. - PubMed
-
- Hong D, Gupta R, Ancliff P, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. - PubMed
-
- Castor A, Nilsson L, Astrand-Grundström I, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med. 2005;11:630–637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

