Exploring RNA structural codes with SHAPE chemistry
- PMID: 21615079
- PMCID: PMC3177967
- DOI: 10.1021/ar200051h
Exploring RNA structural codes with SHAPE chemistry
Abstract
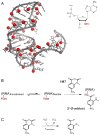
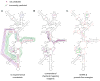
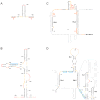
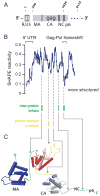
RNA is the central conduit for gene expression. This role depends on an ability to encode information at two levels: in its linear sequence and in the complex structures RNA can form by folding back on itself. Understanding the global structure-function interrelationships mediated by RNA remains a great challenge in molecular and structural biology. In this Account, we discuss evolving work in our laboratory focused on creating facile, generic, quantitative, accurate, and highly informative approaches for understanding RNA structure in biologically important environments. The core innovation derives from our discovery that the nucleophilic reactivity of the ribose 2'-hydroxyl in RNA is gated by local nucleotide flexibility. The 2'-hydroxyl is reactive at conformationally flexible positions but is unreactive at nucleotides constrained by base pairing. Sites of modification in RNA can be detected efficiently either using primer extension or by protection from exoribonucleolytic degradation. This technology is now called SHAPE, for selective 2'-hydroxyl acylation analyzed by primer extension (or protection from exoribonuclease). SHAPE reactivities are largely independent of nucleotide identity but correlate closely with model-free measurements of molecular order. The simple SHAPE reaction is thus a robust, nucleotide-resolution, biophysical measurement of RNA structure. SHAPE can be used to provide an experimental correction to RNA folding algorithms and, in favorable cases, yield kilobase-scale secondary structure predictions with high accuracies. SHAPE chemistry is based on very simple reactive carbonyl centers that can be varied to yield slow- and fast-reacting reagents. Differential SHAPE reactivities can be used to detect specific RNA positions with slow local nucleotide dynamics. These positions, which are often in the C2'-endo conformation, have the potential to function as molecular timers that regulate RNA folding and function. In addition, fast-reacting SHAPE reagents can be used to visualize RNA structural biogenesis and RNA-protein assembly reactions in one second snapshots in very straightforward experiments. The application of SHAPE to challenging problems in biology has revealed surprises in well-studied systems. New regions have been identified that are likely to have critical functional roles on the basis of their high levels of RNA structure. For example, SHAPE analysis of large RNAs, such as authentic viral RNA genomes, suggests that RNA structure organizes regulatory motifs and regulates splicing, protein folding, genome recombination, and ribonucleoprotein assembly. SHAPE has also revealed limitations to the hierarchical model for RNA folding. Continued development and application of SHAPE technologies will advance our understanding of the many ways in which the genetic code is expressed through the underlying structure of RNA.
Figures

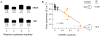



Similar articles
-
RNA structural analysis by evolving SHAPE chemistry.Wiley Interdiscip Rev RNA. 2014 Nov-Dec;5(6):867-81. doi: 10.1002/wrna.1253. Epub 2014 Aug 15. Wiley Interdiscip Rev RNA. 2014. PMID: 25132067 Free PMC article. Review.
-
Slow conformational dynamics at C2'-endo nucleotides in RNA.J Am Chem Soc. 2008 Jul 16;130(28):8884-5. doi: 10.1021/ja802691e. Epub 2008 Jun 18. J Am Chem Soc. 2008. PMID: 18558680 Free PMC article.
-
Selective 2'-hydroxyl acylation analyzed by protection from exoribonuclease (RNase-detected SHAPE) for direct analysis of covalent adducts and of nucleotide flexibility in RNA.Nat Protoc. 2011 Oct 6;6(11):1683-94. doi: 10.1038/nprot.2011.373. Nat Protoc. 2011. PMID: 21979276 Free PMC article.
-
Time-resolved RNA SHAPE chemistry.J Am Chem Soc. 2008 Dec 3;130(48):16178-80. doi: 10.1021/ja8061216. J Am Chem Soc. 2008. PMID: 18998638
-
SHAPE Directed Discovery of New Functions in Large RNAs.Acc Chem Res. 2021 May 18;54(10):2502-2517. doi: 10.1021/acs.accounts.1c00118. Epub 2021 May 7. Acc Chem Res. 2021. PMID: 33960770 Free PMC article. Review.
Cited by
-
Model-Free RNA Sequence and Structure Alignment Informed by SHAPE Probing Reveals a Conserved Alternate Secondary Structure for 16S rRNA.PLoS Comput Biol. 2015 May 20;11(5):e1004126. doi: 10.1371/journal.pcbi.1004126. eCollection 2015 May. PLoS Comput Biol. 2015. PMID: 25992778 Free PMC article.
-
Molecular Dynamics Simulations with Grand-Canonical Reweighting Suggest Cooperativity Effects in RNA Structure Probing Experiments.J Chem Theory Comput. 2023 Jun 27;19(12):3672-3685. doi: 10.1021/acs.jctc.3c00084. Epub 2023 Jun 8. J Chem Theory Comput. 2023. PMID: 37288967 Free PMC article.
-
Restriction of an intron size en route to endothermy.Nucleic Acids Res. 2021 Mar 18;49(5):2460-2487. doi: 10.1093/nar/gkab046. Nucleic Acids Res. 2021. PMID: 33550394 Free PMC article.
-
Mutually exclusive RNA secondary structures regulate translation initiation of DinQ in Escherichia coli.RNA. 2016 Nov;22(11):1739-1749. doi: 10.1261/rna.058461.116. Epub 2016 Sep 20. RNA. 2016. PMID: 27651528 Free PMC article.
-
Computational approaches to predicting the impact of novel bases on RNA structure and stability.ACS Chem Biol. 2013 Nov 15;8(11):2354-9. doi: 10.1021/cb4006062. Epub 2013 Oct 8. ACS Chem Biol. 2013. PMID: 24063428 Free PMC article. Review.
References
-
- Muesing MA, Smith DH, Capon DJ. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. - PubMed
-
- Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. - PubMed
-
- Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci. 2009;34:16–24. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

