The internal Kaposi's sarcoma-associated herpesvirus LANA regions exert a critical role on episome persistence
- PMID: 21593163
- PMCID: PMC3147901
- DOI: 10.1128/JVI.00304-11
The internal Kaposi's sarcoma-associated herpesvirus LANA regions exert a critical role on episome persistence
Abstract
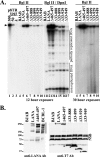
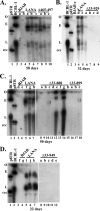
Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen (LANA) is a 1,162-amino-acid protein that acts on viral terminal repeat (TR) DNA to mediate KSHV episome persistence. The two essential components of episome persistence are DNA replication prior to cell division and episome segregation to daughter nuclei. These functions are located within N- and C-terminal regions of LANA. N- and C-terminal regions of LANA are sufficient for TR DNA replication. In addition, N- and C-terminal regions of LANA tether episomes to mitotic chromosomes to segregate episomes to progeny cell nuclei. To generate a tethering mechanism, N-terminal LANA binds histones H2A/H2B to attach to mitotic chromosomes, and C-terminal LANA binds TR DNA and also associates with mitotic chromosomes. Here, we test the importance of the internal LANA sequence for episome persistence. We generated LANA mutants that contain N- and C-terminal regions of LANA but have most of the internal sequence deleted. As expected, the LANA mutants bound mitotic chromosomes in a wild-type pattern and also bound TR DNA as assayed by electrophoretic mobility shift assays (EMSA). The mutants mediated TR DNA replication, although with reduced efficiency compared with LANA. Despite the ability to replicate DNA and exert the chromosome and DNA binding functions necessary for segregating episomes to daughter nuclei, the mutants were highly deficient for the ability to mediate both short- and long-term episome persistence. These data indicate that internal LANA sequence exerts a critical effect on its ability to maintain episomes, possibly through effects on TR DNA replication.
Figures



Similar articles
-
Identification of Kaposi's sarcoma-associated herpesvirus LANA regions important for episome segregation, replication, and persistence.J Virol. 2013 Nov;87(22):12270-83. doi: 10.1128/JVI.01243-13. Epub 2013 Sep 4. J Virol. 2013. PMID: 24006437 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus LANA-Adjacent Regions with Distinct Functions in Episome Segregation or Maintenance.J Virol. 2019 Mar 5;93(6):e02158-18. doi: 10.1128/JVI.02158-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626680 Free PMC article.
-
Role of Kaposi's sarcoma-associated herpesvirus C-terminal LANA chromosome binding in episome persistence.J Virol. 2009 May;83(9):4326-37. doi: 10.1128/JVI.02395-08. Epub 2009 Feb 18. J Virol. 2009. PMID: 19225000 Free PMC article.
-
Kaposi's Sarcoma Herpesvirus Genome Persistence.Front Microbiol. 2016 Aug 12;7:1149. doi: 10.3389/fmicb.2016.01149. eCollection 2016. Front Microbiol. 2016. PMID: 27570517 Free PMC article. Review.
-
KSHV Genome Replication and Maintenance.Front Microbiol. 2016 Feb 1;7:54. doi: 10.3389/fmicb.2016.00054. eCollection 2016. Front Microbiol. 2016. PMID: 26870016 Free PMC article. Review.
Cited by
-
Kaposi's sarcoma-associated herpesvirus suppression of DUSP1 facilitates cellular pathogenesis following de novo infection.J Virol. 2013 Jan;87(1):621-35. doi: 10.1128/JVI.01441-12. Epub 2012 Oct 24. J Virol. 2013. PMID: 23097457 Free PMC article.
-
KSHV LANA acetylation-selective acidic domain reader sequence mediates virus persistence.Proc Natl Acad Sci U S A. 2020 Sep 8;117(36):22443-22451. doi: 10.1073/pnas.2004809117. Epub 2020 Aug 20. Proc Natl Acad Sci U S A. 2020. PMID: 32820070 Free PMC article.
-
Human Gammaherpesvirus 8 Oncogenes Associated with Kaposi's Sarcoma.Int J Mol Sci. 2022 Jun 29;23(13):7203. doi: 10.3390/ijms23137203. Int J Mol Sci. 2022. PMID: 35806208 Free PMC article. Review.
-
A short sequence immediately upstream of the internal repeat elements is critical for KSHV LANA mediated DNA replication and impacts episome persistence.Virology. 2014 Jan 5;448:344-55. doi: 10.1016/j.virol.2013.10.026. Epub 2013 Nov 12. Virology. 2014. PMID: 24314665 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus LANA Modulates the Stability of the E3 Ubiquitin Ligase RLIM.J Virol. 2020 Feb 14;94(5):e01578-19. doi: 10.1128/JVI.01578-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31801865 Free PMC article.
References
-
- An J., Sun Y., Rettig M. B. 2004. Transcriptional coactivation of c-Jun by the KSHV-encoded LANA. Blood 103:222–228 - PubMed
-
- Bajaj B. G., et al. 2006. KSHV encoded LANA upregulates Pim-1 and is a substrate for its kinase activity. Virology 351:18–28 - PubMed
-
- Ballestas M. E., Chatis P. A., Kaye K. M. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641–644 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

