The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid
- PMID: 21593146
- PMCID: PMC3147902
- DOI: 10.1128/JVI.00325-11
The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid
Abstract
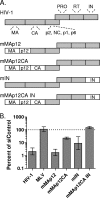
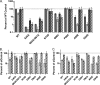
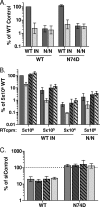
Lentiviruses likely infect nondividing cells by commandeering host nuclear transport factors to facilitate the passage of their preintegration complexes (PICs) through nuclear pore complexes (NPCs) within nuclear envelopes. Genome-wide small interfering RNA screens previously identified karyopherin β transportin-3 (TNPO3) and NPC component nucleoporin 153 (NUP153) as being important for infection by human immunodeficiency virus type 1 (HIV-1). The knockdown of either protein significantly inhibited HIV-1 infectivity, while infection by the gammaretrovirus Moloney murine leukemia virus (MLV) was unaffected. Here, we establish that primate lentiviruses are particularly sensitive to NUP153 knockdown and investigate HIV-1-encoded elements that contribute to this dependency. Mutants lacking functional Vpr or the central DNA flap remained sensitive to NUP153 depletion, while MLV/HIV-1 chimera viruses carrying MLV matrix, capsid, or integrase became less sensitive when the latter two elements were substituted. Two capsid missense mutant viruses, N74D and P90A, were largely insensitive to NUP153 depletion, as was wild-type HIV-1 when cyclophilin A was depleted simultaneously or when infection was conducted in the presence of cyclosporine A. The codepletion of NUP153 and TNPO3 yielded synergistic effects that outweighed those calculated based on individual knockdowns, indicating potential interdependent roles for these factors during HIV-1 infection. Quantitative PCR revealed normal levels of late reverse transcripts, a moderate reduction of 2-long terminal repeat (2-LTR) circles, and a relatively large reduction in integrated proviruses upon NUP153 knockdown. These results suggest that capsid, likely by the qualities of its uncoating, determines whether HIV-1 requires cellular NUP153 for PIC nuclear import.
Figures

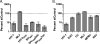
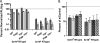
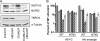

Comment in
-
Examining the requirements for nucleoporins by HIV-1.Future Microbiol. 2011 Nov;6(11):1247-50. doi: 10.2217/fmb.11.111. Future Microbiol. 2011. PMID: 22082286
Similar articles
-
Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells.J Virol. 2018 Sep 12;92(19):e00648-18. doi: 10.1128/JVI.00648-18. Print 2018 Oct 1. J Virol. 2018. PMID: 29997211 Free PMC article.
-
The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase.J Virol. 2010 Jan;84(1):397-406. doi: 10.1128/JVI.01899-09. Epub 2009 Oct 21. J Virol. 2010. PMID: 19846519 Free PMC article.
-
Interplay between HIV entry and transportin-SR2 dependency.Retrovirology. 2011 Jan 30;8:7. doi: 10.1186/1742-4690-8-7. Retrovirology. 2011. PMID: 21276267 Free PMC article.
-
Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes.Viruses. 2013 Oct 7;5(10):2483-511. doi: 10.3390/v5102483. Viruses. 2013. PMID: 24103892 Free PMC article. Review.
-
HIV Capsid and Integration Targeting.Viruses. 2021 Jan 18;13(1):125. doi: 10.3390/v13010125. Viruses. 2021. PMID: 33477441 Free PMC article. Review.
Cited by
-
The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating.J Virol. 2013 Jan;87(1):422-32. doi: 10.1128/JVI.07177-11. Epub 2012 Oct 24. J Virol. 2013. PMID: 23097435 Free PMC article.
-
Defining the HIV Capsid Binding Site of Nucleoporin 153.mSphere. 2022 Oct 26;7(5):e0031022. doi: 10.1128/msphere.00310-22. Epub 2022 Aug 30. mSphere. 2022. PMID: 36040047 Free PMC article.
-
The importance of becoming double-stranded: Innate immunity and the kinetic model of HIV-1 central plus strand synthesis.Virology. 2013 Jun 20;441(1):1-11. doi: 10.1016/j.virol.2013.03.010. Epub 2013 Apr 3. Virology. 2013. PMID: 23561461 Free PMC article. Review.
-
Human immunodeficiency virus type 1 capsid mutation N74D alters cyclophilin A dependence and impairs macrophage infection.J Virol. 2012 Apr;86(8):4708-14. doi: 10.1128/JVI.05887-11. Epub 2012 Feb 1. J Virol. 2012. PMID: 22301145 Free PMC article.
-
3D structure determination of native mammalian cells using cryo-FIB and cryo-electron tomography.J Struct Biol. 2012 Nov;180(2):318-26. doi: 10.1016/j.jsb.2012.07.003. Epub 2012 Jul 14. J Struct Biol. 2012. PMID: 22796867 Free PMC article.
References
-
- Ball J. R., Ullman K. S. 2005. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma 114:319–330 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

