Enhanced polyubiquitination of Shank3 and NMDA receptor in a mouse model of autism
- PMID: 21565394
- PMCID: PMC3110672
- DOI: 10.1016/j.cell.2011.03.052
Enhanced polyubiquitination of Shank3 and NMDA receptor in a mouse model of autism
Retraction in
-
Retraction notice to: Enhanced polyubiquitination of Shank3 and NMDA receptor in a mouse model of autism.Cell. 2013 Jan 17;152(1-2):367. doi: 10.1016/j.cell.2012.12.039. Cell. 2013. PMID: 23332766 Free PMC article. No abstract available.
Abstract
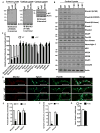
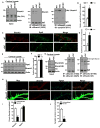
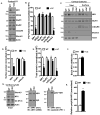
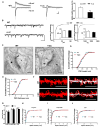
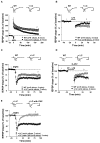
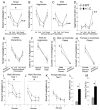
We have created a mouse genetic model that mimics a human mutation of Shank3 that deletes the C terminus and is associated with autism. Expressed as a single copy [Shank3(+/ΔC) mice], Shank3ΔC protein interacts with the wild-type (WT) gene product and results in >90% reduction of Shank3 at synapses. This "gain-of-function" phenotype is linked to increased polyubiquitination of WT Shank3 and its redistribution into proteasomes. Similarly, the NR1 subunit of the NMDA receptor is reduced at synapses with increased polyubiquitination. Assays of postsynaptic density proteins, spine morphology, and synapse number are unchanged in Shank3(+/ΔC) mice, but the amplitude of NMDAR responses is reduced together with reduced NMDAR-dependent LTP and LTD. Reciprocally, mGluR-dependent LTD is markedly enhanced. Shank3(+/ΔC) mice show behavioral deficits suggestive of autism and reduced NMDA receptor function. These studies reveal a mechanism distinct from haploinsufficiency by which mutations of Shank3 can evoke an autism-like disorder.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
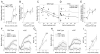
Similar articles
-
Autism-associated Shank3 mutations alter mGluR expression and mGluR-dependent but not NMDA receptor-dependent long-term depression.Synapse. 2019 Aug;73(8):e22097. doi: 10.1002/syn.22097. Epub 2019 Mar 23. Synapse. 2019. PMID: 30868621
-
Early life sleep disruption potentiates lasting sex-specific changes in behavior in genetically vulnerable Shank3 heterozygous autism model mice.Mol Autism. 2022 Aug 29;13(1):35. doi: 10.1186/s13229-022-00514-5. Mol Autism. 2022. PMID: 36038911 Free PMC article.
-
Importance of Shank3 protein in regulating metabotropic glutamate receptor 5 (mGluR5) expression and signaling at synapses.J Biol Chem. 2011 Oct 7;286(40):34839-50. doi: 10.1074/jbc.M111.258384. Epub 2011 Jul 27. J Biol Chem. 2011. PMID: 21795692 Free PMC article.
-
Modeling autism by SHANK gene mutations in mice.Neuron. 2013 Apr 10;78(1):8-27. doi: 10.1016/j.neuron.2013.03.016. Neuron. 2013. PMID: 23583105 Free PMC article. Review.
-
The Role of Zinc and NMDA Receptors in Autism Spectrum Disorders.Pharmaceuticals (Basel). 2022 Dec 20;16(1):1. doi: 10.3390/ph16010001. Pharmaceuticals (Basel). 2022. PMID: 36678498 Free PMC article. Review.
Cited by
-
Neuroligin1 drives synaptic and behavioral maturation through intracellular interactions.J Neurosci. 2013 May 29;33(22):9364-84. doi: 10.1523/JNEUROSCI.4660-12.2013. J Neurosci. 2013. PMID: 23719805 Free PMC article.
-
Knockout of NMDA receptors in parvalbumin interneurons recreates autism-like phenotypes.Autism Res. 2013 Apr;6(2):69-77. doi: 10.1002/aur.1264. Epub 2013 Feb 25. Autism Res. 2013. PMID: 23441094 Free PMC article.
-
The molecular basis of cognitive deficits in pervasive developmental disorders.Learn Mem. 2012 Aug 16;19(9):434-43. doi: 10.1101/lm.025007.111. Learn Mem. 2012. PMID: 22904374 Free PMC article. Review.
-
Protein interactome reveals converging molecular pathways among autism disorders.Sci Transl Med. 2011 Jun 8;3(86):86ra49. doi: 10.1126/scitranslmed.3002166. Sci Transl Med. 2011. PMID: 21653829 Free PMC article.
-
The Notch ligand E3 ligase, Mind Bomb1, regulates glutamate receptor localization in Drosophila.Mol Cell Neurosci. 2016 Jan;70:11-21. doi: 10.1016/j.mcn.2015.11.004. Epub 2015 Nov 17. Mol Cell Neurosci. 2016. PMID: 26596173 Free PMC article.
References
-
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. - PubMed
-
- Beneken J, Tu JC, Xiao B, Nuriya M, Yuan JP, Worley PF, Leahy DJ. Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron. 2000;26:143–154. - PubMed
-
- Brakeman PR, Lanahan AA, O’Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. - PubMed
-
- Condra JA, Neibergs H, Wei W, Brennan MD. Evidence for two schizophrenia susceptibility genes on chromosome 22q13. Psychiatr Genet. 2007;17:292–298. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 MH084020/MH/NIMH NIH HHS/United States
- R21 HD065290-01/HD/NICHD NIH HHS/United States
- AS7098/Autism Speaks/United States
- R01 NS070301-01/NS/NINDS NIH HHS/United States
- R01 MH081164/MH/NIMH NIH HHS/United States
- R01 HD069560-01/HD/NICHD NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 HD069560/HD/NICHD NIH HHS/United States
- R21HD065290/HD/NICHD NIH HHS/United States
- R01MH081164/MH/NIMH NIH HHS/United States
- 5P50MH084020-03/MH/NIMH NIH HHS/United States
- 5R01NS070301-02/NS/NINDS NIH HHS/United States
- R21 HD065290/HD/NICHD NIH HHS/United States
- R01 NS070301-02/NS/NINDS NIH HHS/United States
- R01 NS070301/NS/NINDS NIH HHS/United States
- R01 HD069560-02/HD/NICHD NIH HHS/United States
- R21 HD065290-02/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

