Evolution of the karyopherin-β family of nucleocytoplasmic transport factors; ancient origins and continued specialization
- PMID: 21556326
- PMCID: PMC3083441
- DOI: 10.1371/journal.pone.0019308
Evolution of the karyopherin-β family of nucleocytoplasmic transport factors; ancient origins and continued specialization
Abstract
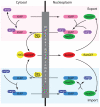
Background: Macromolecular transport across the nuclear envelope (NE) is achieved through nuclear pore complexes (NPCs) and requires karyopherin-βs (KAP-βs), a family of soluble receptors, for recognition of embedded transport signals within cargo. We recently demonstrated, through proteomic analysis of trypanosomes, that NPC architecture is likely highly conserved across the Eukaryota, which in turn suggests conservation of the transport mechanisms. To determine if KAP-β diversity was similarly established early in eukaryotic evolution or if it was subsequently layered onto a conserved NPC, we chose to identify KAP-β sequences in a diverse range of eukaryotes and to investigate their evolutionary history.
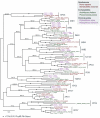
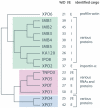
Results: Thirty six predicted proteomes were scanned for candidate KAP-β family members. These resulting sequences were resolved into fifteen KAP-β subfamilies which, due to broad supergroup representation, were most likely represented in the last eukaryotic common ancestor (LECA). Candidate members of each KAP-β subfamily were found in all eukaryotic supergroups, except XPO6, which is absent from Archaeplastida. Phylogenetic reconstruction revealed the likely evolutionary relationships between these different subfamilies. Many species contain more than one representative of each KAP-β subfamily; many duplications are apparently taxon-specific but others result from duplications occurring earlier in eukaryotic history.
Conclusions: At least fifteen KAP-β subfamilies were established early in eukaryote evolution and likely before the LECA. In addition we identified expansions at multiple stages within eukaryote evolution, including a multicellular plant-specific KAP-β, together with frequent secondary losses. Taken with evidence for early establishment of NPC architecture, these data demonstrate that multiple pathways for nucleocytoplasmic transport were established prior to the radiation of modern eukaryotes but that selective pressure continues to sculpt the KAP-β family.
Conflict of interest statement
Figures



Similar articles
-
Karyopherin-mediated nucleocytoplasmic transport.Nat Rev Mol Cell Biol. 2022 May;23(5):307-328. doi: 10.1038/s41580-021-00446-7. Epub 2022 Jan 20. Nat Rev Mol Cell Biol. 2022. PMID: 35058649 Free PMC article. Review.
-
Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor.Mol Cell Proteomics. 2009 Sep;8(9):2119-30. doi: 10.1074/mcp.M900038-MCP200. Epub 2009 Jun 13. Mol Cell Proteomics. 2009. PMID: 19525551 Free PMC article.
-
Karyopherin beta 2B participates in mRNA export from the nucleus.Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14195-9. doi: 10.1073/pnas.212518199. Epub 2002 Oct 16. Proc Natl Acad Sci U S A. 2002. PMID: 12384575 Free PMC article.
-
Rules for nuclear localization sequence recognition by karyopherin beta 2.Cell. 2006 Aug 11;126(3):543-58. doi: 10.1016/j.cell.2006.05.049. Cell. 2006. PMID: 16901787 Free PMC article.
-
Molecular mechanisms of nuclear protein transport.Crit Rev Eukaryot Gene Expr. 1997;7(1-2):61-72. doi: 10.1615/critreveukargeneexpr.v7.i1-2.40. Crit Rev Eukaryot Gene Expr. 1997. PMID: 9034715 Review.
Cited by
-
Multiple types of nuclear localization signals in Entamoeba histolytica.Biochem Biophys Rep. 2024 Jul 4;39:101770. doi: 10.1016/j.bbrep.2024.101770. eCollection 2024 Sep. Biochem Biophys Rep. 2024. PMID: 39055170 Free PMC article.
-
Comparative genomics of nuclear envelope proteins.BMC Genomics. 2018 Nov 16;19(1):823. doi: 10.1186/s12864-018-5218-4. BMC Genomics. 2018. PMID: 30445911 Free PMC article.
-
Touching from a distance.Nucleus. 2014 Jul-Aug;5(4):304-10. doi: 10.4161/nucl.32228. Nucleus. 2014. PMID: 25482119 Free PMC article.
-
Floppy but not sloppy: Interaction mechanism of FG-nucleoporins and nuclear transport receptors.Semin Cell Dev Biol. 2017 Aug;68:34-41. doi: 10.1016/j.semcdb.2017.06.026. Epub 2017 Jun 30. Semin Cell Dev Biol. 2017. PMID: 28669824 Free PMC article. Review.
-
An automated graphics tool for comparative genomics: the Coulson plot generator.BMC Bioinformatics. 2013 Apr 27;14:141. doi: 10.1186/1471-2105-14-141. BMC Bioinformatics. 2013. PMID: 23621955 Free PMC article.
References
-
- Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. - PubMed
-
- Dacks JB, Field MC. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J Cell Sci. 2007;120:2977–2985. - PubMed
-
- Field MC, Dacks JB. First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr Opin Cell Biol. 2009;21:4–13. - PubMed
-
- Mason DA, Stage DE, Goldfarb DS. Evolution of the metazoan-specific importin alpha gene family. J Mol Evol. 2009;68:351–365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

