Hepatitis C virus hijacks P-body and stress granule components around lipid droplets
- PMID: 21543503
- PMCID: PMC3126564
- DOI: 10.1128/JVI.02418-10
Hepatitis C virus hijacks P-body and stress granule components around lipid droplets
Abstract
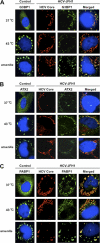
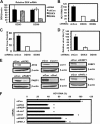
The microRNA miR-122 and DDX6/Rck/p54, a microRNA effector, have been implicated in hepatitis C virus (HCV) replication. In this study, we demonstrated for the first time that HCV-JFH1 infection disrupted processing (P)-body formation of the microRNA effectors DDX6, Lsm1, Xrn1, PATL1, and Ago2, but not the decapping enzyme DCP2, and dynamically redistributed these microRNA effectors to the HCV production factory around lipid droplets in HuH-7-derived RSc cells. Notably, HCV-JFH1 infection also redistributed the stress granule components GTPase-activating protein (SH3 domain)-binding protein 1 (G3BP1), ataxin-2 (ATX2), and poly(A)-binding protein 1 (PABP1) to the HCV production factory. In this regard, we found that the P-body formation of DDX6 began to be disrupted at 36 h postinfection. Consistently, G3BP1 transiently formed stress granules at 36 h postinfection. We then observed the ringlike formation of DDX6 or G3BP1 and colocalization with HCV core after 48 h postinfection, suggesting that the disruption of P-body formation and the hijacking of P-body and stress granule components occur at a late step of HCV infection. Furthermore, HCV infection could suppress stress granule formation in response to heat shock or treatment with arsenite. Importantly, we demonstrate that the accumulation of HCV RNA was significantly suppressed in DDX6, Lsm1, ATX2, and PABP1 knockdown cells after the inoculation of HCV-JFH1, suggesting that the P-body and the stress granule components are required for the HCV life cycle. Altogether, HCV seems to hijack the P-body and the stress granule components for HCV replication.
Figures
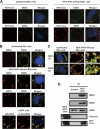
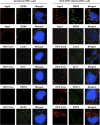
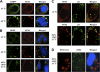
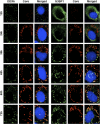

Similar articles
-
Hepatitis C virus infection alters P-body composition but is independent of P-body granules.J Virol. 2012 Aug;86(16):8740-9. doi: 10.1128/JVI.07167-11. Epub 2012 Jun 6. J Virol. 2012. PMID: 22674998 Free PMC article.
-
Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress.J Virol. 2012 Oct;86(20):11043-56. doi: 10.1128/JVI.07101-11. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855484 Free PMC article.
-
Hepatitis C virus co-opts Ras-GTPase-activating protein-binding protein 1 for its genome replication.J Virol. 2011 Jul;85(14):6996-7004. doi: 10.1128/JVI.00013-11. Epub 2011 May 11. J Virol. 2011. PMID: 21561913 Free PMC article.
-
Hepatitis C virus plays with fire and yet avoids getting burned. A review for clinicians on processing bodies and stress granules.Liver Int. 2018 Mar;38(3):388-398. doi: 10.1111/liv.13541. Epub 2017 Sep 4. Liver Int. 2018. PMID: 28782251 Review.
-
Hepatitis C virus utilizes lipid droplet for production of infectious virus.Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(7):217-28. doi: 10.2183/pjab.85.217. Proc Jpn Acad Ser B Phys Biol Sci. 2009. PMID: 19644222 Free PMC article. Review.
Cited by
-
Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses.Virology. 2015 May;479-480:457-74. doi: 10.1016/j.virol.2015.03.001. Epub 2015 Mar 26. Virology. 2015. PMID: 25818028 Free PMC article. Review.
-
MicroRNAs, hepatitis C virus, and HCV/HIV-1 co-infection: new insights in pathogenesis and therapy.Viruses. 2012 Oct 26;4(11):2485-513. doi: 10.3390/v4112485. Viruses. 2012. PMID: 23202492 Free PMC article. Review.
-
Targeting Nup358/RanBP2 by a viral protein disrupts stress granule formation.PLoS Pathog. 2022 Dec 1;18(12):e1010598. doi: 10.1371/journal.ppat.1010598. eCollection 2022 Dec. PLoS Pathog. 2022. PMID: 36455064 Free PMC article.
-
Hepatitis C virus infection restricts human LINE-1 retrotransposition in hepatoma cells.PLoS Pathog. 2021 Apr 19;17(4):e1009496. doi: 10.1371/journal.ppat.1009496. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33872335 Free PMC article.
-
Cytoplasmic hGle1A regulates stress granules by modulation of translation.Mol Biol Cell. 2015 Apr 15;26(8):1476-90. doi: 10.1091/mbc.E14-11-1523. Epub 2015 Feb 18. Mol Biol Cell. 2015. PMID: 25694449 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

