Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS
- PMID: 21541367
- PMCID: PMC3082519
- DOI: 10.1371/journal.pbio.1000614
Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS
Abstract
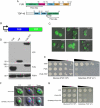
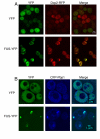
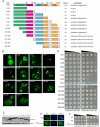
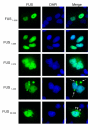
TDP-43 and FUS are RNA-binding proteins that form cytoplasmic inclusions in some forms of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). Moreover, mutations in TDP-43 and FUS are linked to ALS and FTLD. However, it is unknown whether TDP-43 and FUS aggregate and cause toxicity by similar mechanisms. Here, we exploit a yeast model and purified FUS to elucidate mechanisms of FUS aggregation and toxicity. Like TDP-43, FUS must aggregate in the cytoplasm and bind RNA to confer toxicity in yeast. These cytoplasmic FUS aggregates partition to stress granule compartments just as they do in ALS patients. Importantly, in isolation, FUS spontaneously forms pore-like oligomers and filamentous structures reminiscent of FUS inclusions in ALS patients. FUS aggregation and toxicity requires a prion-like domain, but unlike TDP-43, additional determinants within a RGG domain are critical for FUS aggregation and toxicity. In further distinction to TDP-43, ALS-linked FUS mutations do not promote aggregation. Finally, genome-wide screens uncovered stress granule assembly and RNA metabolism genes that modify FUS toxicity but not TDP-43 toxicity. Our findings suggest that TDP-43 and FUS, though similar RNA-binding proteins, aggregate and confer disease phenotypes via distinct mechanisms. These differences will likely have important therapeutic implications.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
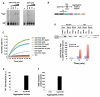
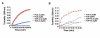
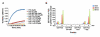

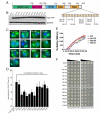

Comment in
-
A yeast model for understanding ALS: fast, cheap, and easy to control.PLoS Biol. 2011 Apr;9(4):e1001053. doi: 10.1371/journal.pbio.1001053. Epub 2011 Apr 26. PLoS Biol. 2011. PMID: 21541366 Free PMC article. No abstract available.
Similar articles
-
RNA-binding proteins with prion-like domains in ALS and FTLD-U.Prion. 2011 Jul-Sep;5(3):179-87. doi: 10.4161/pri.5.3.17230. Epub 2011 Jul 1. Prion. 2011. PMID: 21847013 Free PMC article. Review.
-
How do the RNA-binding proteins TDP-43 and FUS relate to amyotrophic lateral sclerosis and frontotemporal degeneration, and to each other?Curr Opin Neurol. 2012 Dec;25(6):701-7. doi: 10.1097/WCO.0b013e32835a269b. Curr Opin Neurol. 2012. PMID: 23041957 Review.
-
Conjoint pathologic cascades mediated by ALS/FTLD-U linked RNA-binding proteins TDP-43 and FUS.Neurology. 2011 Oct 25;77(17):1636-43. doi: 10.1212/WNL.0b013e3182343365. Epub 2011 Sep 28. Neurology. 2011. PMID: 21956718 Free PMC article. Review.
-
FUS/TLS forms cytoplasmic aggregates, inhibits cell growth and interacts with TDP-43 in a yeast model of amyotrophic lateral sclerosis.Protein Cell. 2011 Mar;2(3):223-36. doi: 10.1007/s13238-011-1525-0. Epub 2011 Mar 30. Protein Cell. 2011. PMID: 21452073 Free PMC article.
-
The RNA-binding motif 45 (RBM45) protein accumulates in inclusion bodies in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) patients.Acta Neuropathol. 2012 Nov;124(5):717-32. doi: 10.1007/s00401-012-1045-x. Epub 2012 Sep 21. Acta Neuropathol. 2012. PMID: 22993125 Free PMC article.
Cited by
-
Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis.Brain Res. 2016 Sep 15;1647:65-78. doi: 10.1016/j.brainres.2016.03.036. Epub 2016 Mar 28. Brain Res. 2016. PMID: 27033831 Free PMC article. Review.
-
Inside out: the role of nucleocytoplasmic transport in ALS and FTLD.Acta Neuropathol. 2016 Aug;132(2):159-173. doi: 10.1007/s00401-016-1586-5. Epub 2016 Jun 6. Acta Neuropathol. 2016. PMID: 27271576 Free PMC article. Review.
-
Drivers of Hsp104 potentiation revealed by scanning mutagenesis of the middle domain.Protein Sci. 2021 Aug;30(8):1667-1685. doi: 10.1002/pro.4126. Epub 2021 Jun 1. Protein Sci. 2021. PMID: 34010483 Free PMC article.
-
Expression of Fused in sarcoma mutations in mice recapitulates the neuropathology of FUS proteinopathies and provides insight into disease pathogenesis.Mol Neurodegener. 2012 Oct 10;7:53. doi: 10.1186/1750-1326-7-53. Mol Neurodegener. 2012. PMID: 23046583 Free PMC article.
-
FUS Microphase Separation: Regulation by Nucleic Acid Polymers and DNA Repair Proteins.Int J Mol Sci. 2022 Oct 30;23(21):13200. doi: 10.3390/ijms232113200. Int J Mol Sci. 2022. PMID: 36361989 Free PMC article.
References
-
- Rosen D, Siddique T, Patterson D, Figlewicz D, Sapp P, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. - PubMed
-
- Cleveland D. W, Rothstein J. D. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. - PubMed
-
- Neumann M, Sampathu D. M, Kwong L. K, Truax A. C, Micsenyi M. C, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. - PubMed
-
- Kwong L. K, Neumann M, Sampathu D. M, Lee V. M, Trojanowski J. Q. TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathol. 2007;114:63–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

