The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella
- PMID: 21525242
- PMCID: PMC3128531
- DOI: 10.1091/mbc.E10-11-0893
The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella
Abstract
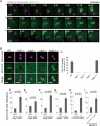
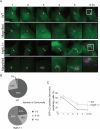
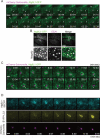
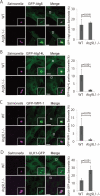
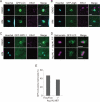
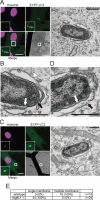
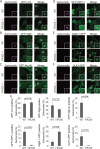
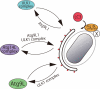
Salmonella develops into resident bacteria in epithelial cells, and the autophagic machinery (Atg) is thought to play an important role in this process. In this paper, we show that an autophagosome-like double-membrane structure surrounds the Salmonella still residing within the Salmonella-containing vacuole (SCV). This double membrane is defective in Atg9L1- and FAK family-interacting protein of 200 kDa (FIP200)-deficient cells. Atg9L1 and FIP200 are important for autophagy-specific recruitment of the phosphatidylinositol 3-kinase (PI3K) complex. However, in the absence of Atg9L1, FIP200, and the PI3K complex, LC3 and its E3-like enzyme, the Atg16L complex, are still recruited to Salmonella. We propose that the LC3 system is recruited through a mechanism that is independent of isolation membrane generation.
Figures
Similar articles
-
Three-Axis Model for Atg Recruitment in Autophagy against Salmonella.Int J Cell Biol. 2012;2012:389562. doi: 10.1155/2012/389562. Epub 2012 Feb 28. Int J Cell Biol. 2012. PMID: 22505927 Free PMC article.
-
Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages.PLoS Pathog. 2014 Jun 5;10(6):e1004159. doi: 10.1371/journal.ppat.1004159. eCollection 2014 Jun. PLoS Pathog. 2014. PMID: 24901456 Free PMC article.
-
Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17?Autophagy. 2009 Jan;5(1):85-7. doi: 10.4161/auto.5.1.7180. Epub 2009 Jan 13. Autophagy. 2009. PMID: 18981720
-
Molecular basis of canonical and bactericidal autophagy.Int Immunol. 2009 Nov;21(11):1199-204. doi: 10.1093/intimm/dxp088. Epub 2009 Sep 7. Int Immunol. 2009. PMID: 19737785 Review.
-
Autophagy basics.Microbiol Immunol. 2011 Jan;55(1):1-11. doi: 10.1111/j.1348-0421.2010.00271.x. Microbiol Immunol. 2011. PMID: 21175768 Review.
Cited by
-
The ULK1 complex: sensing nutrient signals for autophagy activation.Autophagy. 2013 Feb 1;9(2):124-37. doi: 10.4161/auto.23323. Epub 2013 Jan 7. Autophagy. 2013. PMID: 23295650 Free PMC article. Review.
-
Autophagy proteins in macroendocytic engulfment.Trends Cell Biol. 2012 Jul;22(7):374-80. doi: 10.1016/j.tcb.2012.04.005. Epub 2012 May 19. Trends Cell Biol. 2012. PMID: 22608991 Free PMC article. Review.
-
The incredible ULKs.Cell Commun Signal. 2012 Mar 13;10(1):7. doi: 10.1186/1478-811X-10-7. Cell Commun Signal. 2012. PMID: 22413737 Free PMC article.
-
WIPI-1 Positive Autophagosome-Like Vesicles Entrap Pathogenic Staphylococcus aureus for Lysosomal Degradation.Int J Cell Biol. 2012;2012:179207. doi: 10.1155/2012/179207. Epub 2012 Jul 9. Int J Cell Biol. 2012. PMID: 22829830 Free PMC article.
-
LC3-Associated Phagocytosis in Bacterial Infection.Pathogens. 2022 Jul 30;11(8):863. doi: 10.3390/pathogens11080863. Pathogens. 2022. PMID: 36014984 Free PMC article. Review.
References
-
- Bakowski MA, Braun V, Brumell JH. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic. 2008;9:2022–2031. - PubMed
-
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

