DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis
- PMID: 21521783
- PMCID: PMC3128482
- DOI: 10.1182/blood-2011-02-334359
DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis
Abstract
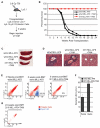
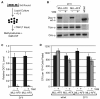
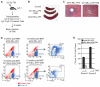
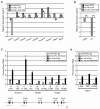
Chromosomal translocations of the mixed lineage leukemia (MLL) gene are a common cause of acute leukemias. The oncogenic function of MLL fusion proteins is, in part, mediated through aberrant activation of Hoxa genes and Meis1, among others. Here we demonstrate using a tamoxifen-inducible Cre-mediated loss of function mouse model that DOT1L, an H3K79 methyltransferase, is required for both initiation and maintenance of MLL-AF9-induced leukemogenesis in vitro and in vivo. Through gene expression and chromatin immunoprecipitation analysis we demonstrate that mistargeting of DOT1L, subsequent H3K79 methylation, and up-regulation of Hoxa and Meis1 genes underlie the molecular mechanism of how DOT1L contributes to MLL-AF9-mediated leukemogenesis. Our study not only provides the first in vivo evidence for the function of DOT1L in leukemia, but also reveals the molecular mechanism for DOT1L in MLL-AF9 mediated leukemia. Thus, DOT1L may serve as a potential therapeutic target for the treatment of leukemia caused by MLL translocations.
Figures


Similar articles
-
MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L.Cancer Cell. 2011 Jul 12;20(1):66-78. doi: 10.1016/j.ccr.2011.06.010. Cancer Cell. 2011. PMID: 21741597 Free PMC article.
-
Targeting recruitment of disruptor of telomeric silencing 1-like (DOT1L): characterizing the interactions between DOT1L and mixed lineage leukemia (MLL) fusion proteins.J Biol Chem. 2013 Oct 18;288(42):30585-30596. doi: 10.1074/jbc.M113.457135. Epub 2013 Sep 1. J Biol Chem. 2013. PMID: 23996074 Free PMC article.
-
Degree of recruitment of DOT1L to MLL-AF9 defines level of H3K79 Di- and tri-methylation on target genes and transformation potential.Cell Rep. 2015 May 5;11(5):808-20. doi: 10.1016/j.celrep.2015.04.004. Epub 2015 Apr 23. Cell Rep. 2015. PMID: 25921540 Free PMC article.
-
Targeting DOT1L and HOX gene expression in MLL-rearranged leukemia and beyond.Exp Hematol. 2015 Aug;43(8):673-84. doi: 10.1016/j.exphem.2015.05.012. Epub 2015 Jun 25. Exp Hematol. 2015. PMID: 26118503 Free PMC article. Review.
-
The role of DOT1L in the maintenance of leukemia gene expression.Curr Opin Genet Dev. 2016 Feb;36:68-72. doi: 10.1016/j.gde.2016.03.015. Epub 2016 May 3. Curr Opin Genet Dev. 2016. PMID: 27151433 Review.
Cited by
-
Perturbation of Methionine/S-adenosylmethionine Metabolism as a Novel Vulnerability in MLL Rearranged Leukemia.Cells. 2019 Oct 25;8(11):1322. doi: 10.3390/cells8111322. Cells. 2019. PMID: 31717699 Free PMC article.
-
The fusion partner specifies the oncogenic potential of NUP98 fusion proteins.Leuk Res. 2013 Dec;37(12):1668-73. doi: 10.1016/j.leukres.2013.09.013. Epub 2013 Sep 21. Leuk Res. 2013. PMID: 24090997 Free PMC article.
-
Identification of DOT1L inhibitors by structure-based virtual screening adapted from a nucleoside-focused library.Eur J Med Chem. 2020 Mar 1;189:112023. doi: 10.1016/j.ejmech.2019.112023. Epub 2020 Jan 2. Eur J Med Chem. 2020. PMID: 31978781 Free PMC article.
-
Epigenetic regulation of protein translation in KMT2A-rearranged AML.Exp Hematol. 2020 May;85:57-69. doi: 10.1016/j.exphem.2020.04.007. Epub 2020 May 11. Exp Hematol. 2020. PMID: 32437908 Free PMC article.
-
Therapeutic strategies against hDOT1L as a potential drug target in MLL-rearranged leukemias.Clin Epigenetics. 2020 May 25;12(1):73. doi: 10.1186/s13148-020-00860-2. Clin Epigenetics. 2020. PMID: 32450905 Free PMC article. Review.
References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. - PubMed
-
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6(11):838–849. - PubMed
-
- Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115(3):293–303. - PubMed
-
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20(40):5695–5707. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

