Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells
- PMID: 21514197
- PMCID: PMC3099128
- DOI: 10.1016/j.molcel.2011.04.005
Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells
Abstract
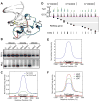
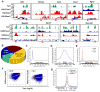
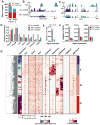
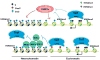
DNA methylation at the 5 position of cytosine (5mC) in the mammalian genome is a key epigenetic event critical for various cellular processes. The ten-eleven translocation (Tet) family of 5mC-hydroxylases, which convert 5mC to 5-hydroxymethylcytosine (5hmC), offers a way for dynamic regulation of DNA methylation. Here we report that Tet1 binds to unmodified C or 5mC- or 5hmC-modified CpG-rich DNA through its CXXC domain. Genome-wide mapping of Tet1 and 5hmC reveals mechanisms by which Tet1 controls 5hmC and 5mC levels in mouse embryonic stem cells (mESCs). We also uncover a comprehensive gene network influenced by Tet1. Collectively, our data suggest that Tet1 controls DNA methylation both by binding to CpG-rich regions to prevent unwanted DNA methyltransferase activity, and by converting 5mC to 5hmC through hydroxylase activity. This Tet1-mediated antagonism of CpG methylation imparts differential maintenance of DNA methylation status at Tet1 targets, ultimately contributing to mESC differentiation and the onset of embryonic development.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Tet1 and 5-hydroxymethylation: a genome-wide view in mouse embryonic stem cells.Cell Cycle. 2011 Aug 1;10(15):2428-36. doi: 10.4161/cc.10.15.16930. Epub 2011 Aug 1. Cell Cycle. 2011. PMID: 21750410 Free PMC article.
-
MYC deregulates TET1 and TET2 expression to control global DNA (hydroxy)methylation and gene expression to maintain a neoplastic phenotype in T-ALL.Epigenetics Chromatin. 2019 Jul 2;12(1):41. doi: 10.1186/s13072-019-0278-5. Epigenetics Chromatin. 2019. PMID: 31266538 Free PMC article.
-
Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells.Genes Dev. 2011 Apr 1;25(7):679-84. doi: 10.1101/gad.2036011. Genes Dev. 2011. PMID: 21460036 Free PMC article.
-
Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation.Genes Dev. 2011 Dec 1;25(23):2436-52. doi: 10.1101/gad.179184.111. Genes Dev. 2011. PMID: 22156206 Free PMC article. Review.
-
Role of ten-eleven translocation proteins and 5-hydroxymethylcytosine in hepatocellular carcinoma.Cell Prolif. 2019 Jul;52(4):e12626. doi: 10.1111/cpr.12626. Epub 2019 Apr 29. Cell Prolif. 2019. PMID: 31033072 Free PMC article. Review.
Cited by
-
The role of DNA methylation in aging, rejuvenation, and age-related disease.Rejuvenation Res. 2012 Oct;15(5):483-94. doi: 10.1089/rej.2012.1324. Rejuvenation Res. 2012. PMID: 23098078 Free PMC article. Review.
-
Active DNA Demethylase, TET1, Increases Oxidative Phosphorylation and Sensitizes Ovarian Cancer Stem Cells to Mitochondrial Complex I Inhibitor.Antioxidants (Basel). 2024 Jun 17;13(6):735. doi: 10.3390/antiox13060735. Antioxidants (Basel). 2024. PMID: 38929174 Free PMC article.
-
miR-29 regulates Tet1 expression and contributes to early differentiation of mouse ESCs.Oncotarget. 2016 Oct 4;7(40):64932-64941. doi: 10.18632/oncotarget.10751. Oncotarget. 2016. PMID: 27449105 Free PMC article.
-
Impact of retrotransposons in pluripotent stem cells.Mol Cells. 2012 Dec;34(6):509-16. doi: 10.1007/s10059-012-0242-8. Epub 2012 Nov 6. Mol Cells. 2012. PMID: 23135636 Free PMC article. Review.
-
Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency.Nat Cell Biol. 2013 Jun;15(6):700-11. doi: 10.1038/ncb2748. Epub 2013 May 19. Nat Cell Biol. 2013. PMID: 23685628 Free PMC article.
References
-
- Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16:455–462. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

