Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling
- PMID: 21512159
- PMCID: PMC3134964
- DOI: 10.1161/CIRCRESAHA.110.235531
Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling
Abstract
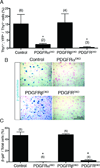
Rationale: In early heart development, platelet-derived growth factor (PDGF) receptor expression in the heart ventricles is restricted to the epicardium. Previously, we showed that PDGFRβ is required for coronary vascular smooth muscle cell (cVSMC) development, but a role for PDGFRα has not been identified. Therefore, we investigated the combined and independent roles of these receptors in epicardial development.
Objective: To understand the contribution of PDGF receptors in epicardial development and epicardial-derived cell fate determination.
Methods and results: By generating mice with epicardial-specific deletion of the PDGF receptors, we found that epicardial epithelial-to-mesenchymal transition (EMT) was defective. Sox9, an SRY-related transcription factor, was reduced in PDGF receptor-deficient epicardial cells, and overexpression of Sox9 restored epicardial migration, actin reorganization, and EMT gene expression profiles. The failure of epicardial EMT resulted in hearts that lacked epicardial-derived cardiac fibroblasts and cVSMC. Loss of PDGFRα resulted in a specific disruption of cardiac fibroblast development, whereas cVSMC development was unperturbed.
Conclusions: Signaling through both PDGF receptors is necessary for epicardial EMT and formation of epicardial-mesenchymal derivatives. PDGF receptors also have independent functions in the development of specific epicardial-derived cell fates.
Figures
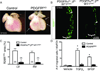
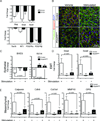

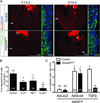
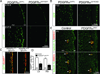

Similar articles
-
Nf1 limits epicardial derivative expansion by regulating epithelial to mesenchymal transition and proliferation.Development. 2012 Jun;139(11):2040-9. doi: 10.1242/dev.074054. Epub 2012 Apr 25. Development. 2012. PMID: 22535408 Free PMC article.
-
Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations.Circ Res. 2008 Dec 5;103(12):1393-401. doi: 10.1161/CIRCRESAHA.108.176768. Epub 2008 Oct 23. Circ Res. 2008. PMID: 18948621 Free PMC article.
-
In vitro epithelial-to-mesenchymal transformation in human adult epicardial cells is regulated by TGFβ-signaling and WT1.Basic Res Cardiol. 2011 Sep;106(5):829-47. doi: 10.1007/s00395-011-0181-0. Epub 2011 Apr 24. Basic Res Cardiol. 2011. PMID: 21516490 Free PMC article.
-
Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease.Circ Res. 2012 Jun 8;110(12):1628-45. doi: 10.1161/CIRCRESAHA.111.259960. Circ Res. 2012. PMID: 22679138 Free PMC article. Review.
-
PDGF-C and PDGF-D signaling in vascular diseases and animal models.Mol Aspects Med. 2018 Aug;62:1-11. doi: 10.1016/j.mam.2018.01.005. Epub 2018 Feb 14. Mol Aspects Med. 2018. PMID: 29410092 Review.
Cited by
-
Novel therapeutic strategies targeting fibroblasts and fibrosis in heart disease.Nat Rev Drug Discov. 2016 Sep;15(9):620-638. doi: 10.1038/nrd.2016.89. Epub 2016 Jun 24. Nat Rev Drug Discov. 2016. PMID: 27339799 Free PMC article. Review.
-
Tcf21 regulates the specification and maturation of proepicardial cells.Development. 2013 Jun;140(11):2409-21. doi: 10.1242/dev.093385. Epub 2013 May 1. Development. 2013. PMID: 23637334 Free PMC article.
-
Hypoxia induced the differentiation of Tbx18-positive epicardial cells to CoSMCs.Sci Rep. 2016 Jul 26;6:30468. doi: 10.1038/srep30468. Sci Rep. 2016. PMID: 27456656 Free PMC article.
-
Twist factor regulation of non-cardiomyocyte cell lineages in the developing heart.Differentiation. 2012 Jul;84(1):79-88. doi: 10.1016/j.diff.2012.03.002. Epub 2012 Apr 17. Differentiation. 2012. PMID: 22516205 Free PMC article. Review.
-
Transcriptional Profiling of Cultured, Embryonic Epicardial Cells Identifies Novel Genes and Signaling Pathways Regulated by TGFβR3 In Vitro.PLoS One. 2016 Aug 9;11(8):e0159710. doi: 10.1371/journal.pone.0159710. eCollection 2016. PLoS One. 2016. PMID: 27505173 Free PMC article.
References
-
- Mellgren AM, Smith CL, Olsen GS, Eskiocak B, Zhou B, Kazi MN, Ruiz FR, Pu WT, Tallquist MD. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ Res. 2008;103:1393–1401. - PMC - PubMed
-
- Kang J, Gu Y, Li P, Johnson BL, Sucov HM, Thomas PS. Pdgf-a as an epicardial mitogen during heart development. Dev Dyn. 2008;237:692–701. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 HL100401-02/HL/NHLBI NIH HHS/United States
- U01 HL100401-01/HL/NHLBI NIH HHS/United States
- T32 HL007360-32/HL/NHLBI NIH HHS/United States
- R01 HL074257-07/HL/NHLBI NIH HHS/United States
- T32 HL007360/HL/NHLBI NIH HHS/United States
- R01 HL074257/HL/NHLBI NIH HHS/United States
- F30 HL096277/HL/NHLBI NIH HHS/United States
- R01 HL074257-02/HL/NHLBI NIH HHS/United States
- R01 HL074257-03/HL/NHLBI NIH HHS/United States
- R01 HL074257-06/HL/NHLBI NIH HHS/United States
- U01 HL100401/HL/NHLBI NIH HHS/United States
- 1F30HL096277-01A1/HL/NHLBI NIH HHS/United States
- R01 HL074257-04/HL/NHLBI NIH HHS/United States
- HL100401/HL/NHLBI NIH HHS/United States
- HL074257/HL/NHLBI NIH HHS/United States
- R01 HL074257-05A2/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

