MiD49 and MiD51, new components of the mitochondrial fission machinery
- PMID: 21508961
- PMCID: PMC3128275
- DOI: 10.1038/embor.2011.54
MiD49 and MiD51, new components of the mitochondrial fission machinery
Abstract
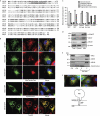
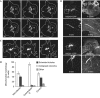
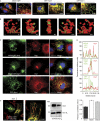
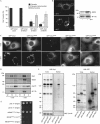
Mitochondria form intricate networks through fission and fusion events. Here, we identify mitochondrial dynamics proteins of 49 and 51 kDa (MiD49 and MiD51, respectively) anchored in the mitochondrial outer membrane. MiD49/51 form foci and rings around mitochondria similar to the fission mediator dynamin-related protein 1 (Drp1). MiD49/51 directly recruit Drp1 to the mitochondrial surface, whereas their knockdown reduces Drp1 association, leading to unopposed fusion. Overexpression of MiD49/51 seems to sequester Drp1 from functioning at mitochondria and cause fused tubules to associate with actin. Thus, MiD49/51 are new mediators of mitochondrial division affecting Drp1 action at mitochondria.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission.J Biol Chem. 2013 Sep 20;288(38):27584-27593. doi: 10.1074/jbc.M113.479873. Epub 2013 Aug 6. J Biol Chem. 2013. PMID: 23921378 Free PMC article.
-
Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission.Mol Biol Cell. 2013 Mar;24(5):659-67. doi: 10.1091/mbc.E12-10-0721. Epub 2013 Jan 2. Mol Biol Cell. 2013. PMID: 23283981 Free PMC article.
-
The role of Drp1 adaptor proteins MiD49 and MiD51 in mitochondrial fission: implications for human disease.Clin Sci (Lond). 2016 Nov 1;130(21):1861-74. doi: 10.1042/CS20160030. Clin Sci (Lond). 2016. PMID: 27660309 Review.
-
Loss of MIEF1/MiD51 confers susceptibility to BAX-mediated cell death and PINK1-PRKN-dependent mitophagy.Autophagy. 2019 Dec;15(12):2107-2125. doi: 10.1080/15548627.2019.1596494. Epub 2019 Mar 28. Autophagy. 2019. PMID: 30894073 Free PMC article.
-
Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1.Ann N Y Acad Sci. 2010 Jul;1201:34-9. doi: 10.1111/j.1749-6632.2010.05629.x. Ann N Y Acad Sci. 2010. PMID: 20649536 Free PMC article. Review.
Cited by
-
Mitochondrial Quality Control Orchestrates the Symphony of B Cells and Plays Critical Roles in B Cell-Related Diseases.J Immunol Res. 2024 Oct 17;2024:5577506. doi: 10.1155/2024/5577506. eCollection 2024. J Immunol Res. 2024. PMID: 39449998 Free PMC article. Review.
-
Molecular mechanisms of mitochondrial dynamics.Nat Rev Mol Cell Biol. 2024 Oct 17. doi: 10.1038/s41580-024-00785-1. Online ahead of print. Nat Rev Mol Cell Biol. 2024. PMID: 39420231 Review.
-
SUMOylation of MFF coordinates fission complexes to promote stress-induced mitochondrial fragmentation.Sci Adv. 2024 Oct 4;10(40):eadq6223. doi: 10.1126/sciadv.adq6223. Epub 2024 Oct 4. Sci Adv. 2024. PMID: 39365854 Free PMC article.
-
Role of Mitochondrial Dysfunctions in Neurodegenerative Disorders: Advances in Mitochondrial Biology.Mol Neurobiol. 2024 Sep 13. doi: 10.1007/s12035-024-04469-x. Online ahead of print. Mol Neurobiol. 2024. PMID: 39269547 Review.
-
The Potential of Mitochondrial Therapeutics in the Treatment of Oxidative Stress and Inflammation in Aging.Mol Neurobiol. 2024 Sep 4. doi: 10.1007/s12035-024-04474-0. Online ahead of print. Mol Neurobiol. 2024. PMID: 39230868 Review.
References
-
- Anesti V, Scorrano L (2006) The relationship between mitochondrial shape and function and the cytoskeleton. Biochim Biophys Acta 1757: 692–699 - PubMed
-
- Bhar D, Karren MA, Babst M, Shaw JM (2006) Dimeric Dnm1–G385D interacts with Mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J Biol Chem 281: 17312–17320 - PubMed
-
- Chang CR, Blackstone C (2007) Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem 282: 21583–21587 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

