Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques
- PMID: 21490952
- PMCID: PMC3072369
- DOI: 10.1371/journal.ppat.1002015
Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques
Abstract
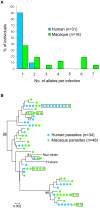
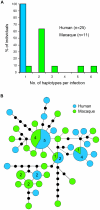
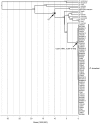
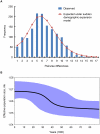
Plasmodium knowlesi, a malaria parasite originally thought to be restricted to macaques in Southeast Asia, has recently been recognized as a significant cause of human malaria. Unlike the benign and morphologically similar P. malariae, these parasites can lead to fatal infections. Malaria parasites, including P. knowlesi, have not yet been detected in macaques of the Kapit Division of Malaysian Borneo, where the majority of human knowlesi malaria cases have been reported. In order to extend our understanding of the epidemiology and evolutionary history of P. knowlesi, we examined 108 wild macaques for malaria parasites and sequenced the circumsporozoite protein (csp) gene and mitochondrial (mt) DNA of P. knowlesi isolates derived from macaques and humans. We detected five species of Plasmodium (P. knowlesi, P. inui, P. cynomolgi, P. fieldi and P. coatneyi) in the long-tailed and pig-tailed macaques, and an extremely high prevalence of P. inui and P. knowlesi. Macaques had a higher number of P. knowlesi genotypes per infection than humans, and some diverse alleles of the P. knowlesi csp gene and certain mtDNA haplotypes were shared between both hosts. Analyses of DNA sequence data indicate that there are no mtDNA lineages associated exclusively with either host. Furthermore, our analyses of the mtDNA data reveal that P. knowlesi is derived from an ancestral parasite population that existed prior to human settlement in Southeast Asia, and underwent significant population expansion approximately 30,000-40,000 years ago. Our results indicate that human infections with P. knowlesi are not newly emergent in Southeast Asia and that knowlesi malaria is primarily a zoonosis with wild macaques as the reservoir hosts. However, ongoing ecological changes resulting from deforestation, with an associated increase in the human population, could enable this pathogenic species of Plasmodium to switch to humans as the preferred host.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Malaria parasites in macaques in Thailand: stump-tailed macaques (Macaca arctoides) are new natural hosts for Plasmodium knowlesi, Plasmodium inui, Plasmodium coatneyi and Plasmodium fieldi.Malar J. 2020 Oct 1;19(1):350. doi: 10.1186/s12936-020-03424-0. Malar J. 2020. PMID: 33004070 Free PMC article.
-
Prevalence of simian malaria parasites in macaques of Singapore.PLoS Negl Trop Dis. 2021 Jan 25;15(1):e0009110. doi: 10.1371/journal.pntd.0009110. eCollection 2021 Jan. PLoS Negl Trop Dis. 2021. PMID: 33493205 Free PMC article.
-
Distribution and prevalence of malaria parasites among long-tailed macaques (Macaca fascicularis) in regional populations across Southeast Asia.Malar J. 2016 Sep 2;15(1):450. doi: 10.1186/s12936-016-1494-0. Malar J. 2016. PMID: 27590474 Free PMC article.
-
Human infections and detection of Plasmodium knowlesi.Clin Microbiol Rev. 2013 Apr;26(2):165-84. doi: 10.1128/CMR.00079-12. Clin Microbiol Rev. 2013. PMID: 23554413 Free PMC article. Review.
-
The Role of Ecological Linkage Mechanisms in Plasmodium knowlesi Transmission and Spread.Ecohealth. 2019 Dec;16(4):594-610. doi: 10.1007/s10393-019-01395-6. Epub 2019 Jan 23. Ecohealth. 2019. PMID: 30675676 Review.
Cited by
-
Evaluation of Mosquito Magnet and other collection tools for Anopheles mosquito vectors of simian malaria.Parasit Vectors. 2021 Apr 1;14(1):184. doi: 10.1186/s13071-021-04689-3. Parasit Vectors. 2021. PMID: 33794965 Free PMC article.
-
Zoonotic pathogens in wild Asian primates: a systematic review highlighting research gaps.Front Vet Sci. 2024 Jun 27;11:1386180. doi: 10.3389/fvets.2024.1386180. eCollection 2024. Front Vet Sci. 2024. PMID: 38993279 Free PMC article.
-
Efficient Surveillance of Plasmodium knowlesi Genetic Subpopulations, Malaysian Borneo, 2000-2018.Emerg Infect Dis. 2020 Jul;26(7):1392-1398. doi: 10.3201/eid2607.190924. Emerg Infect Dis. 2020. PMID: 32568035 Free PMC article.
-
The origin of malarial parasites in orangutans.PLoS One. 2012;7(4):e34990. doi: 10.1371/journal.pone.0034990. Epub 2012 Apr 20. PLoS One. 2012. PMID: 22536346 Free PMC article.
-
Is there evidence of sustained human-mosquito-human transmission of the zoonotic malaria Plasmodium knowlesi? A systematic literature review.Malar J. 2022 Mar 17;21(1):89. doi: 10.1186/s12936-022-04110-z. Malar J. 2022. PMID: 35300703 Free PMC article.
References
-
- Singh B, Sung LK, Matusop A, Radhakrishnan A, Shamsul SSG, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

