The University of Minnesota Pathway Prediction System: multi-level prediction and visualization
- PMID: 21486753
- PMCID: PMC3125723
- DOI: 10.1093/nar/gkr200
The University of Minnesota Pathway Prediction System: multi-level prediction and visualization
Abstract
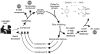
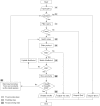
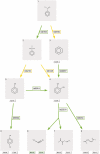
The University of Minnesota Pathway Prediction System (UM-PPS, http://umbbd.msi.umn.edu/predict/) is a rule-based system that predicts microbial catabolism of organic compounds. Currently, its knowledge base contains 250 biotransformation rules and five types of metabolic logic entities. The original UM-PPS predicted up to two prediction levels at a time. Users had to choose a predicted product to continue the prediction. This approach provided a limited view of prediction results and heavily relied on manual intervention. The new UM-PPS produces a multi-level prediction within an acceptable time frame, and allows users to view prediction alternatives much more easily as a directed acyclic graph.
Figures

Similar articles
-
The University of Minnesota pathway prediction system: predicting metabolic logic.Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W427-32. doi: 10.1093/nar/gkn315. Epub 2008 Jun 4. Nucleic Acids Res. 2008. PMID: 18524801 Free PMC article.
-
The University of Minnesota Biocatalysis/Biodegradation Database: the first decade.Nucleic Acids Res. 2006 Jan 1;34(Database issue):D517-21. doi: 10.1093/nar/gkj076. Nucleic Acids Res. 2006. PMID: 16381924 Free PMC article.
-
Microbial pathway prediction: a functional group approach.J Chem Inf Comput Sci. 2003 May-Jun;43(3):1051-7. doi: 10.1021/ci034018f. J Chem Inf Comput Sci. 2003. PMID: 12767164
-
Predicting biodegradation.Environ Microbiol. 1999 Apr;1(2):119-24. Environ Microbiol. 1999. PMID: 11207727 Review.
-
Biotransformation pathway maps in WikiPathways enable direct visualization of drug metabolism related expression changes.Drug Discov Today. 2010 Oct;15(19-20):851-8. doi: 10.1016/j.drudis.2010.08.002. Epub 2010 Aug 11. Drug Discov Today. 2010. PMID: 20708095 Review.
Cited by
-
Computational Reconstruction of NFκB Pathway Interaction Mechanisms during Prostate Cancer.PLoS Comput Biol. 2016 Apr 14;12(4):e1004820. doi: 10.1371/journal.pcbi.1004820. eCollection 2016 Apr. PLoS Comput Biol. 2016. PMID: 27078000 Free PMC article.
-
Omics Approaches to Pesticide Biodegradation.Curr Microbiol. 2020 Apr;77(4):545-563. doi: 10.1007/s00284-020-01916-5. Epub 2020 Feb 20. Curr Microbiol. 2020. PMID: 32078006 Review.
-
Computational Chemical Synthesis Analysis and Pathway Design.Front Chem. 2018 Jun 5;6:199. doi: 10.3389/fchem.2018.00199. eCollection 2018. Front Chem. 2018. PMID: 29915783 Free PMC article. Review.
-
Integration of bioinformatics to biodegradation.Biol Proced Online. 2014 Apr 27;16:8. doi: 10.1186/1480-9222-16-8. eCollection 2014. Biol Proced Online. 2014. PMID: 24808763 Free PMC article. Review.
-
Supervised de novo reconstruction of metabolic pathways from metabolome-scale compound sets.Bioinformatics. 2013 Jul 1;29(13):i135-44. doi: 10.1093/bioinformatics/btt244. Bioinformatics. 2013. PMID: 23812977 Free PMC article.
References
-
- Fenner K, Gao J, Kramer S, Ellis LBM, Wackett LP. Data-driven extraction of relative reasoning rules to limit combinatorial explosion in biodegradation pathway prediction. Bioinformatics. 2008;24:2079–2085. - PubMed
-
- Marchant CA, Briggs KA, Long A. In silico tools for sharing data and knowledge on toxicity and metabolism: derek for windows, meteor, and vitic. Toxicol. Mech. Methods. 2008;18:177–187. - PubMed
-
- Dimitrov S, Nedelcheva D, Dimitrova N, Mekenyan O. Development of a biodegradation model for the prediction of metabolites in soil. Sci. Total Environ. 2010;408:3811–3816. - PubMed

