Structural basis for ligand recognition and discrimination of a quorum-quenching antibody
- PMID: 21454495
- PMCID: PMC3089576
- DOI: 10.1074/jbc.M111.231258
Structural basis for ligand recognition and discrimination of a quorum-quenching antibody
Abstract
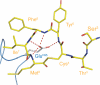
In the postantibiotic era, available treatment options for severe bacterial infections caused by methicillin-resistant Staphylococcus aureus have become limited. Therefore, new and innovative approaches are needed to combat such life-threatening infections. Virulence factor expression in S. aureus is regulated in a cell density-dependent manner using "quorum sensing," which involves generation and secretion of autoinducing peptides (AIPs) into the surrounding environment to activate a bacterial sensor kinase at a particular threshold concentration. Mouse monoclonal antibody AP4-24H11 was shown previously to blunt quorum sensing-mediated changes in gene expression in vitro and protect mice from a lethal dose of S. aureus by sequestering the AIP signal. We have elucidated the crystal structure of the AP4-24H11 Fab in complex with AIP-4 at 2.5 Å resolution to determine its mechanism of ligand recognition. A key Glu(H95) provides much of the binding specificity through formation of hydrogen bonds with each of the four amide nitrogens in the AIP-4 macrocyclic ring. Importantly, these structural data give clues as to the interactions between the cognate staphylococcal AIP receptors AgrC and the AIPs, as AP4-24H11·AIP-4 binding recapitulates features that have been proposed for AgrC-AIP recognition. Additionally, these structural insights may enable the engineering of AIP cross-reactive antibodies or quorum quenching vaccines for use in active or passive immunotherapy for prevention or treatment of S. aureus infections.
Figures
Similar articles
-
Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC.J Mol Biol. 2008 Aug 29;381(2):300-9. doi: 10.1016/j.jmb.2008.06.018. Epub 2008 Jun 13. J Mol Biol. 2008. PMID: 18582472
-
Structural characterization of native autoinducing peptides and abiotic analogues reveals key features essential for activation and inhibition of an AgrC quorum sensing receptor in Staphylococcus aureus.J Am Chem Soc. 2013 Dec 11;135(49):18436-44. doi: 10.1021/ja407533e. Epub 2013 Nov 25. J Am Chem Soc. 2013. PMID: 24219181 Free PMC article.
-
Timing Is Everything: Impact of Naturally Occurring Staphylococcus aureus AgrC Cytoplasmic Domain Adaptive Mutations on Autoinduction.J Bacteriol. 2019 Sep 20;201(20):e00409-19. doi: 10.1128/JB.00409-19. Print 2019 Oct 15. J Bacteriol. 2019. PMID: 31358609 Free PMC article.
-
Quorum-sensing, intra- and inter-species competition in the staphylococci.Microbiology (Reading). 2023 Aug;169(8):001381. doi: 10.1099/mic.0.001381. Microbiology (Reading). 2023. PMID: 37578829 Free PMC article. Review.
-
Deciphering the dynamics of methicillin-resistant Staphylococcus aureus biofilm formation: from molecular signaling to nanotherapeutic advances.Cell Commun Signal. 2024 Mar 22;22(1):188. doi: 10.1186/s12964-024-01511-2. Cell Commun Signal. 2024. PMID: 38519959 Free PMC article. Review.
Cited by
-
Characterization of MroQ-Dependent Maturation and Export of the Staphylococcus aureus Accessory Gene Regulatory System Autoinducing Peptide.Infect Immun. 2022 Oct 20;90(10):e0026322. doi: 10.1128/iai.00263-22. Epub 2022 Sep 8. Infect Immun. 2022. PMID: 36073934 Free PMC article.
-
Conformational Switch to a β-Turn in a Staphylococcal Quorum Sensing Signal Peptide Causes a Dramatic Increase in Potency.J Am Chem Soc. 2020 Jan 15;142(2):750-761. doi: 10.1021/jacs.9b05513. Epub 2020 Jan 2. J Am Chem Soc. 2020. PMID: 31859506 Free PMC article.
-
Highly Stable, Amide-Bridged Autoinducing Peptide Analogues that Strongly Inhibit the AgrC Quorum Sensing Receptor in Staphylococcus aureus.Angew Chem Int Ed Engl. 2016 Jul 25;55(31):8913-7. doi: 10.1002/anie.201602974. Epub 2016 Jun 8. Angew Chem Int Ed Engl. 2016. PMID: 27276693 Free PMC article.
-
Staphylococcus aureus pathogenesis in diverse host environments.Pathog Dis. 2017 Jan 1;75(1):ftx005. doi: 10.1093/femspd/ftx005. Pathog Dis. 2017. PMID: 28104617 Free PMC article. Review.
-
Antibody-Mediated Catalysis in Infection and Immunity.Infect Immun. 2017 Aug 18;85(9):e00202-17. doi: 10.1128/IAI.00202-17. Print 2017 Sep. Infect Immun. 2017. PMID: 28674034 Free PMC article. Review.
References
-
- Reading N. C., Sperandio V. (2006) FEMS. Microbiol. Lett. 254, 1–11 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

