T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1
- PMID: 21448133
- PMCID: PMC3101995
- DOI: 10.1038/emboj.2011.85
T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1
Abstract
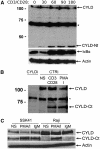
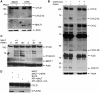
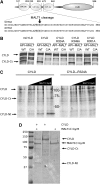
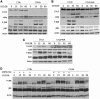
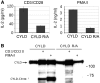
The paracaspase mucosa-associated lymphoid tissue 1 (MALT1) is central to lymphocyte activation and lymphomagenesis. MALT1 mediates antigen receptor signalling to NF-κB by acting as a scaffold protein. Furthermore, MALT1 has proteolytic activity that contributes to optimal NF-κB activation by cleaving the NF-κB inhibitor A20. Whether MALT1 protease activity is involved in other signalling pathways, and the identity of the relevant substrates, is unknown. Here, we show that T-cell receptors (TCR) activation, as well as overexpression of the oncogenic API2-MALT1 fusion protein, results in proteolytic inactivation of CYLD by MALT1, which is specifically required for c-jun N-terminal kinase (JNK) activation and the inducible expression of a subset of genes. These results indicate a novel role for MALT1 proteolytic activity in TCR-induced JNK activation and reveal CYLD cleavage as the underlying mechanism.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20.Nat Immunol. 2008 Mar;9(3):263-71. doi: 10.1038/ni1561. Epub 2008 Jan 27. Nat Immunol. 2008. PMID: 18223652
-
MALT1-dependent cleavage of CYLD promotes NF-κB signaling and growth of aggressive B-cell receptor-dependent lymphomas.Blood Cancer J. 2023 Mar 15;13(1):37. doi: 10.1038/s41408-023-00809-7. Blood Cancer J. 2023. PMID: 36922488 Free PMC article.
-
Protein kinase C-δ negatively regulates T cell receptor-induced NF-κB activation by inhibiting the assembly of CARMA1 signalosome.J Biol Chem. 2012 Jun 8;287(24):20081-7. doi: 10.1074/jbc.M111.335463. Epub 2012 Apr 23. J Biol Chem. 2012. PMID: 22528498 Free PMC article.
-
MALT1 substrate cleavage: what is it good for?Front Immunol. 2024 May 28;15:1412347. doi: 10.3389/fimmu.2024.1412347. eCollection 2024. Front Immunol. 2024. PMID: 38863711 Free PMC article. Review.
-
MALT1--a universal soldier: multiple strategies to ensure NF-κB activation and target gene expression.FEBS J. 2015 Sep;282(17):3286-97. doi: 10.1111/febs.13325. Epub 2015 Jun 10. FEBS J. 2015. PMID: 25996250 Review.
Cited by
-
The subversion of toll-like receptor signaling by bacterial and viral proteases during the development of infectious diseases.Mol Aspects Med. 2022 Dec;88:101143. doi: 10.1016/j.mam.2022.101143. Epub 2022 Sep 21. Mol Aspects Med. 2022. PMID: 36152458 Free PMC article. Review.
-
Mucosa‑associated lymphoid tissue lymphoma translocation protein 1 inhibitor, MI‑2, attenuates non‑small cell lung cancer cell proliferation, migration and invasion, and promotes apoptosis by suppressing the JNK/c‑JUN pathway.Oncol Lett. 2024 Jul 30;28(4):465. doi: 10.3892/ol.2024.14598. eCollection 2024 Oct. Oncol Lett. 2024. PMID: 39119234 Free PMC article.
-
PKCθ/β and CYLD are antagonistic partners in the NFκB and NFAT transactivation pathways in primary mouse CD3+ T lymphocytes.PLoS One. 2013;8(1):e53709. doi: 10.1371/journal.pone.0053709. Epub 2013 Jan 15. PLoS One. 2013. PMID: 23335970 Free PMC article.
-
A MALT1 inhibitor suppresses human myeloid DC, effector T-cell and B-cell responses and retains Th1/regulatory T-cell homeostasis.PLoS One. 2020 Sep 1;15(9):e0222548. doi: 10.1371/journal.pone.0222548. eCollection 2020. PLoS One. 2020. PMID: 32870913 Free PMC article.
-
ADAP regulates cell cycle progression of T cells via control of cyclin E and Cdk2 expression through two distinct CARMA1-dependent signaling pathways.Mol Cell Biol. 2012 May;32(10):1908-17. doi: 10.1128/MCB.06541-11. Epub 2012 Mar 12. Mol Cell Biol. 2012. PMID: 22411628 Free PMC article.
References
-
- Angel P, Hattori K, Smeal T, Karin M (1988) The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell 55: 875–885 - PubMed
-
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R (2008) T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-κB inhibitor A20. Nat Immunol 9: 263–271 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

